|
|
|
|
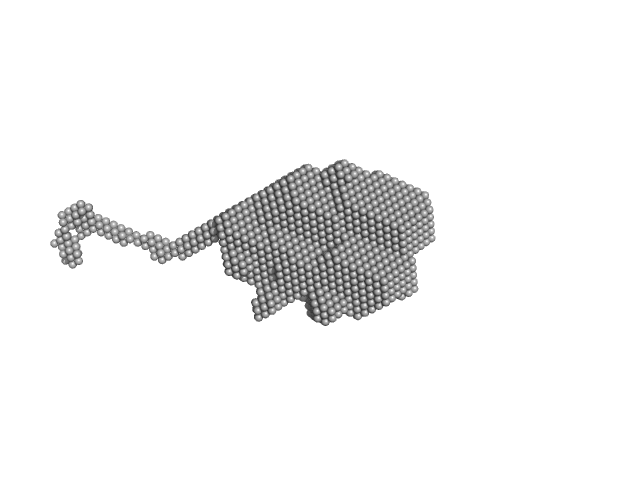
|
| Sample: |
Protein DPCD monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
Deciphering cellular and molecular determinants of human DPCD protein in complex with RUVBL1/RUVBL2 AAA-ATPases.
J Mol Biol :167760 (2022)
Dos Santos Morais R, Santo PE, Ley M, Schelcher C, Abel Y, Plassart L, Deslignière E, Chagot ME, Quinternet M, Paiva ACF, Hessmann S, Morellet N, M F Sousa P, Vandermoere F, Bertrand E, Charpentier B, Bandeiras TM, Plisson-Chastang C, Verheggen C, Cianférani S, Manival X
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RuvB-like 1 hexamer, 311 kDa Homo sapiens protein
RuvB-like 2 hexamer, 311 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
Deciphering cellular and molecular determinants of human DPCD protein in complex with RUVBL1/RUVBL2 AAA-ATPases.
J Mol Biol :167760 (2022)
Dos Santos Morais R, Santo PE, Ley M, Schelcher C, Abel Y, Plassart L, Deslignière E, Chagot ME, Quinternet M, Paiva ACF, Hessmann S, Morellet N, M F Sousa P, Vandermoere F, Bertrand E, Charpentier B, Bandeiras TM, Plisson-Chastang C, Verheggen C, Cianférani S, Manival X
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
1490 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Protein DPCD trimer, 71 kDa Homo sapiens protein
RuvB-like 1 trimer, 155 kDa Homo sapiens protein
RuvB-like 2 trimer, 156 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
Deciphering cellular and molecular determinants of human DPCD protein in complex with RUVBL1/RUVBL2 AAA-ATPases.
J Mol Biol :167760 (2022)
Dos Santos Morais R, Santo PE, Ley M, Schelcher C, Abel Y, Plassart L, Deslignière E, Chagot ME, Quinternet M, Paiva ACF, Hessmann S, Morellet N, M F Sousa P, Vandermoere F, Bertrand E, Charpentier B, Bandeiras TM, Plisson-Chastang C, Verheggen C, Cianférani S, Manival X
|
| RgGuinier |
5.4 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
856 |
nm3 |
|
|
|
|
|
|
|
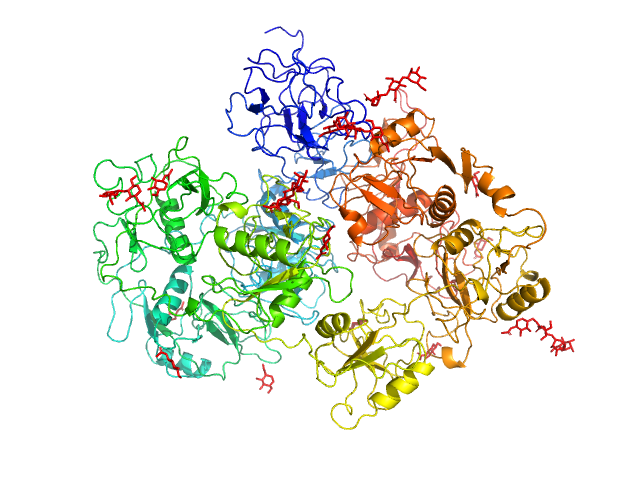
| Sample: |
Secretory phospholipase A2 receptor monomer, 189 kDa Homo sapiens protein
|
| Buffer: |
10 mM Bis-Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 19
|
Structure of PLA2R reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch
Proceedings of the National Academy of Sciences 119(29) (2022)
Fresquet M, Lockhart-Cairns M, Rhoden S, Jowitt T, Briggs D, Baldock C, Brenchley P, Lennon R
|
| RgGuinier |
5.9 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
504 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Secretory phospholipase A2 receptor monomer, 189 kDa Homo sapiens protein
|
| Buffer: |
10 mM Bis-Tris, 150 mM NaCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 19
|
Structure of PLA2R reveals presentation of the dominant membranous nephropathy epitope and an immunogenic patch
Proceedings of the National Academy of Sciences 119(29) (2022)
Fresquet M, Lockhart-Cairns M, Rhoden S, Jowitt T, Briggs D, Baldock C, Brenchley P, Lennon R
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
426 |
nm3 |
|
|
|
|
|
|
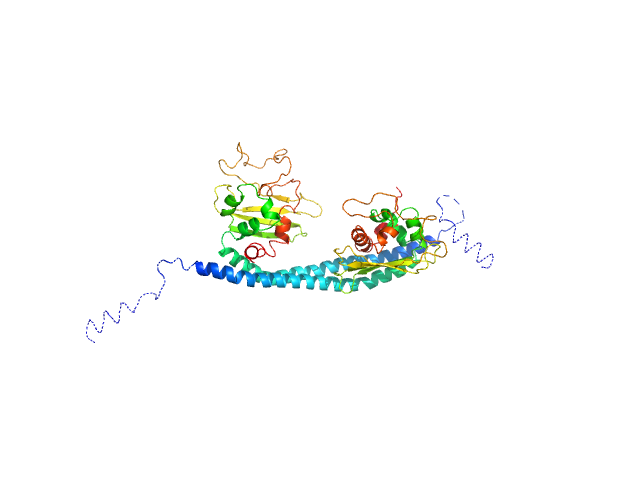
|
| Sample: |
SET nuclear proto-oncogene dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 6.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 11
|
PP2A is activated by cytochrome c upon formation of a diffuse encounter complex with SET/TAF-Iβ
Computational and Structural Biotechnology Journal (2022)
Casado-Combreras M, Rivero-Rodríguez F, Elena-Real C, Molodenskiy D, Díaz-Quintana A, Martinho M, Gerbaud G, González-Arzola K, Velázquez-Campoy A, Svergun D, Belle V, De la Rosa M, Díaz-Moreno I
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
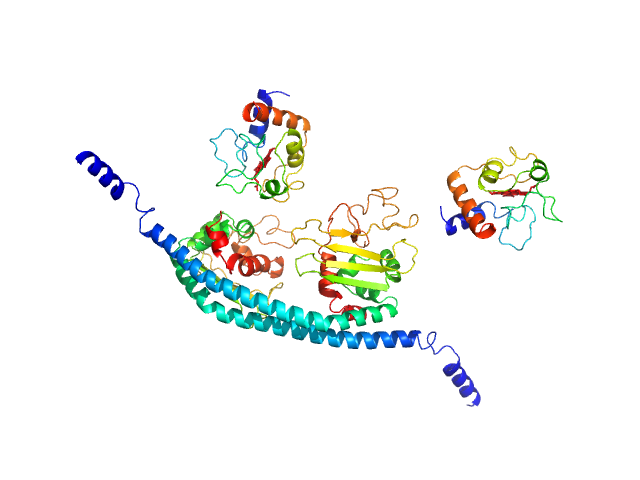
|
| Sample: |
Cytochrome c dimer, 24 kDa Homo sapiens protein
SET nuclear proto-oncogene dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 6.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 11
|
PP2A is activated by cytochrome c upon formation of a diffuse encounter complex with SET/TAF-Iβ
Computational and Structural Biotechnology Journal (2022)
Casado-Combreras M, Rivero-Rodríguez F, Elena-Real C, Molodenskiy D, Díaz-Quintana A, Martinho M, Gerbaud G, González-Arzola K, Velázquez-Campoy A, Svergun D, Belle V, De la Rosa M, Díaz-Moreno I
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
|
|
|
|
|
|
|
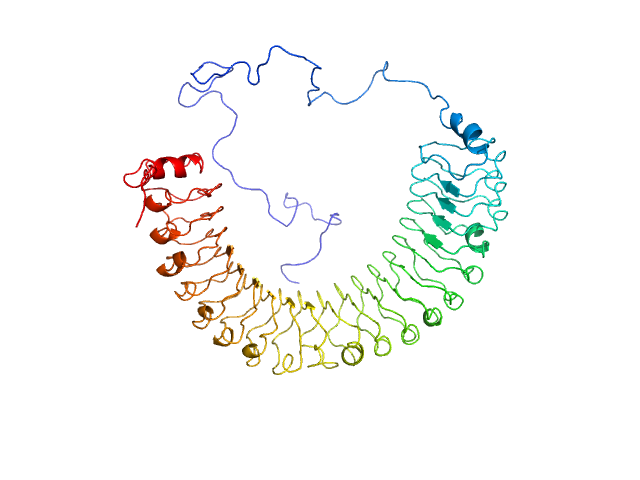
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jun 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
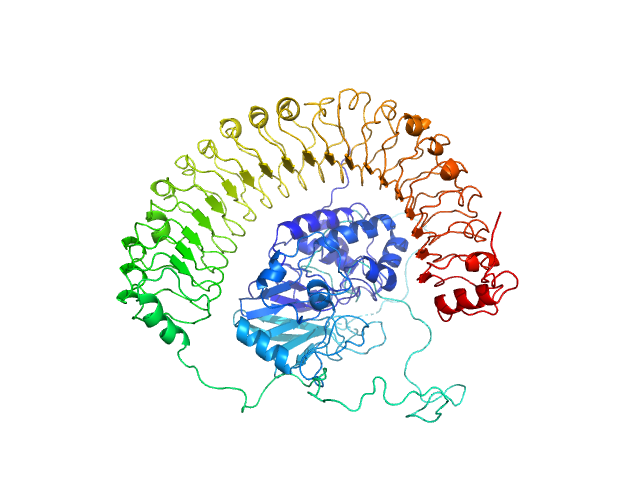
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
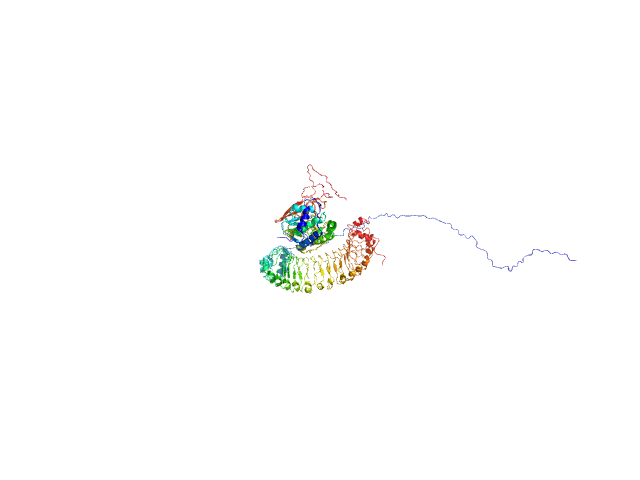
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit monomer, 37 kDa Homo sapiens protein
GTPase KRas monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
156 |
nm3 |
|
|