|
|
|
|
|
| Sample: |
MYC22-G14T/G23T monomer, 7 kDa Homo sapiens DNA
|
| Buffer: |
50 mM Tris, 30 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Feb 27
|
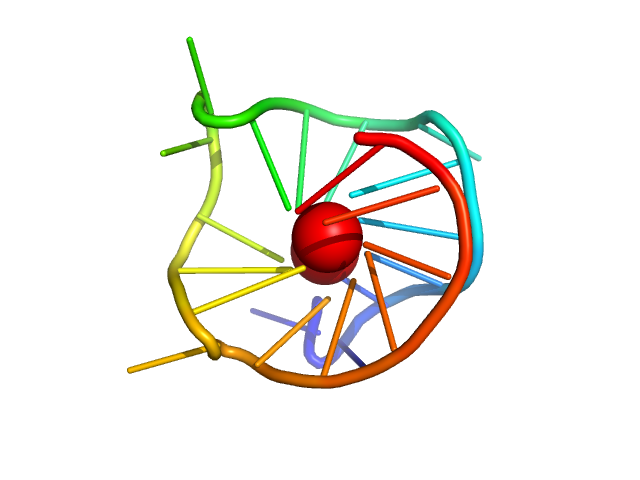
Small-angle X-ray scattering data of a guanine-rich DNA derived from the promoter region of c-MYC gene in solution
Data in Brief :108285 (2022)
Miyauchi K, Imamura H, Yamaoki Y, Kato M
|
| RgGuinier |
1.3 |
nm |
| Dmax |
5.3 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
MYC22-G14T/G23T monomer, 7 kDa Homo sapiens DNA
|
| Buffer: |
50 mM Tris, 100 mM 18-crown-6, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2022 Feb 27
|
Small-angle X-ray scattering data of a guanine-rich DNA derived from the promoter region of c-MYC gene in solution
Data in Brief :108285 (2022)
Miyauchi K, Imamura H, Yamaoki Y, Kato M
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Presequence protease, mitochondrial monomer, 115 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 4
|
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications 13(1) (2022)
Liang W, Wijaya J, Wei H, Noble A, Mancl J, Mo S, Lee D, Lin King J, Pan M, Liu C, Koehler C, Zhao M, Potter C, Carragher B, Li S, Tang W
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Presequence protease, mitochondrial monomer, 115 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 7
|
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications 13(1) (2022)
Liang W, Wijaya J, Wei H, Noble A, Mancl J, Mo S, Lee D, Lin King J, Pan M, Liu C, Koehler C, Zhao M, Potter C, Carragher B, Li S, Tang W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Presequence protease, mitochondrial monomer, 115 kDa Homo sapiens protein
Citrate synthase, mitochondrial monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl , 20 mM EDTA, pH: 7.7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 4
|
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications 13(1) (2022)
Liang W, Wijaya J, Wei H, Noble A, Mancl J, Mo S, Lee D, Lin King J, Pan M, Liu C, Koehler C, Zhao M, Potter C, Carragher B, Li S, Tang W
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Presequence protease, mitochondrial monomer, 115 kDa Homo sapiens protein
Amyloid-beta precursor protein monomer, 4 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl , 20 mM EDTA, pH: 7.7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 7
|
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications 13(1) (2022)
Liang W, Wijaya J, Wei H, Noble A, Mancl J, Mo S, Lee D, Lin King J, Pan M, Liu C, Koehler C, Zhao M, Potter C, Carragher B, Li S, Tang W
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cone-rod homeobox protein monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate,100 mM NaCl, and 5 mM imidazole, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Jun 15
|
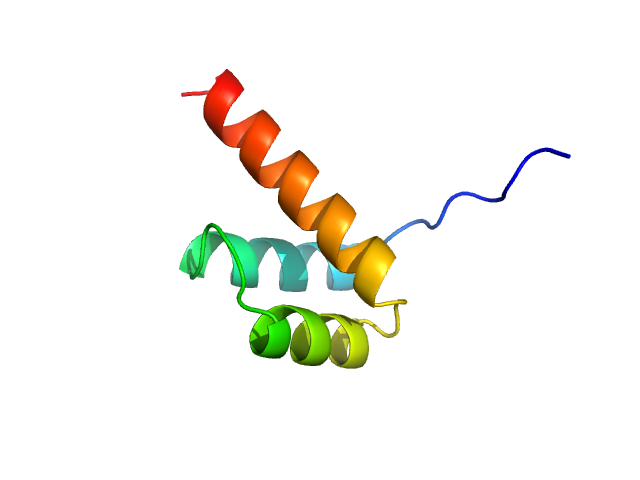
Structural and functional analysis of the human Cone-rod homeobox transcription factor.
Proteins (2022)
Clanor PB, Buchholz C, Hayes JE, Friedman MA, White AM, Enke RA, Berndsen CE
|
| RgGuinier |
1.2 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CUB domain-containing protein 1 monomer, 77 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 137 mM NaCl, 2.7 mM KCl,, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 29
|
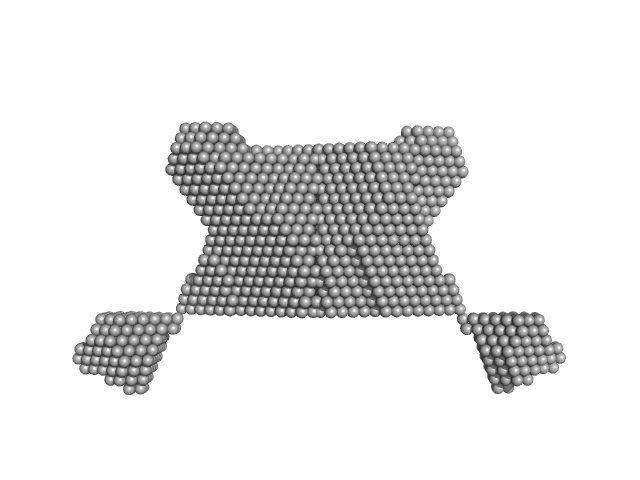
Targeting a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) for RAS-driven cancers.
J Clin Invest 132(4) (2022)
Lim SA, Zhou J, Martinko AJ, Wang YH, Filippova EV, Steri V, Wang D, Remesh SG, Liu J, Hann B, Kossiakoff AA, Evans MJ, Leung KK, Wells JA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
185 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cleaved - CUB domain-containing protein 1 (N-terminus) monomer, 38 kDa Homo sapiens protein
Cleaved - CUB domain-containing protein 1 (C-terminus) monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 137 mM NaCl, 2.7 mM KCl,, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 29
|
Targeting a proteolytic neoepitope on CUB domain containing protein 1 (CDCP1) for RAS-driven cancers.
J Clin Invest 132(4) (2022)
Lim SA, Zhou J, Martinko AJ, Wang YH, Filippova EV, Steri V, Wang D, Remesh SG, Liu J, Hann B, Kossiakoff AA, Evans MJ, Leung KK, Wells JA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
182 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nuclear receptor subfamily 1 group I member 2 monomer, 39 kDa Homo sapiens protein
Nuclear receptor subfamily 1 group I member 3 monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
25 mM Hepes, 150 mM NaCl, 5% glycerol, 5 mM DTT, pH: 7.9 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2021 Oct 8
|
Molecular basis of crosstalk in nuclear receptors: heterodimerization between PXR and CAR and the implication in gene regulation
Shirish chodankar
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|