|
|
|
|

|
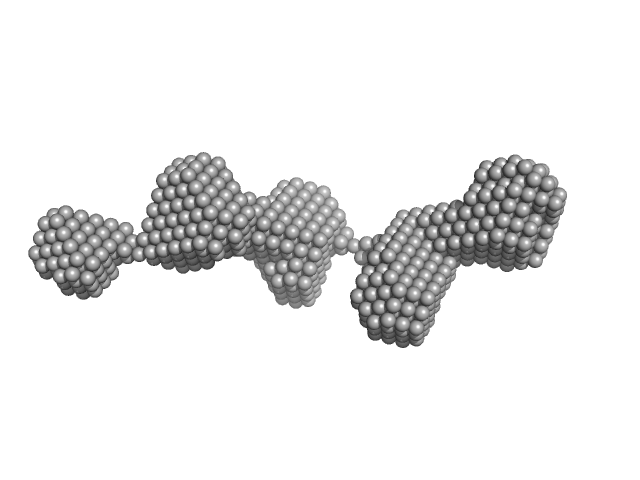
| Sample: |
Fibrillin-1 PF3 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Dec 12
|
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
5.4 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
141 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Latent-transforming growth factor beta-binding protein 1 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Dec 12
|
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Latent-transforming growth factor beta-binding protein 1 monomer, 44 kDa Homo sapiens protein
Fibrillin-1 PF3 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
10 mM Hepes, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 20
|
Latent TGFβ complexes are transglutaminase cross-linked to fibrillin to facilitate TGFβ activation.
Matrix Biol (2022)
Lockhart-Cairns MP, Cain SA, Dajani R, Steer R, Thomson J, Alanazi YF, Kielty CM, Baldock C
|
| RgGuinier |
7.4 |
nm |
| Dmax |
29.6 |
nm |
| VolumePorod |
759 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Teneurin-4 dimer, 537 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Teneurin4 dimer structures reveal a calcium‐stabilized compact conformation supporting homomeric trans‐interactions
The EMBO Journal (2022)
Meijer D, Frias C, Beugelink J, Deurloo Y, Janssen B
|
| RgGuinier |
7.8 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1280 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Teneurin-4 dimer, 537 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Teneurin4 dimer structures reveal a calcium‐stabilized compact conformation supporting homomeric trans‐interactions
The EMBO Journal (2022)
Meijer D, Frias C, Beugelink J, Deurloo Y, Janssen B
|
| RgGuinier |
7.2 |
nm |
| Dmax |
29.5 |
nm |
| VolumePorod |
1270 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Teneurin-4 dimer, 537 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM EDTA, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Teneurin4 dimer structures reveal a calcium‐stabilized compact conformation supporting homomeric trans‐interactions
The EMBO Journal (2022)
Meijer D, Frias C, Beugelink J, Deurloo Y, Janssen B
|
| RgGuinier |
9.5 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
1370 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Teneurin-4 (S2585C) dimer, 537 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Teneurin4 dimer structures reveal a calcium‐stabilized compact conformation supporting homomeric trans‐interactions
The EMBO Journal (2022)
Meijer D, Frias C, Beugelink J, Deurloo Y, Janssen B
|
| RgGuinier |
7.1 |
nm |
| Dmax |
29.5 |
nm |
| VolumePorod |
1260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Teneurin-4 (S2585C) dimer, 537 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Teneurin4 dimer structures reveal a calcium‐stabilized compact conformation supporting homomeric trans‐interactions
The EMBO Journal (2022)
Meijer D, Frias C, Beugelink J, Deurloo Y, Janssen B
|
| RgGuinier |
7.0 |
nm |
| Dmax |
29.5 |
nm |
| VolumePorod |
1270 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Teneurin-4 (S2585C) dimer, 537 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM EDTA, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 16
|
Teneurin4 dimer structures reveal a calcium‐stabilized compact conformation supporting homomeric trans‐interactions
The EMBO Journal (2022)
Meijer D, Frias C, Beugelink J, Deurloo Y, Janssen B
|
| RgGuinier |
7.8 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1390 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Unconventional myosin-X component dimer, 15 kDa Homo sapiens protein
|
| Buffer: |
HEPES, 5% glycerol, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 Mar 16
|
K
-edge anomalous SAXS for protein solution structure modeling
Acta Crystallographica Section D Structural Biology 78(2) (2022)
Virk K, Yonezawa K, Choukate K, Singh L, Shimizu N, Chaudhuri B
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
24 |
nm3 |
|
|