|
|
|
|
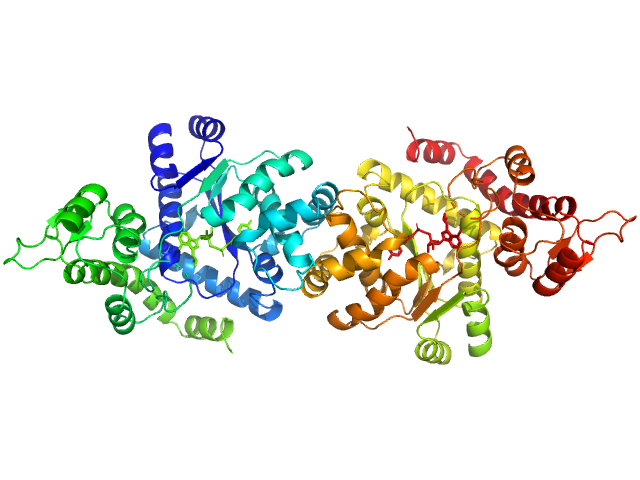
|
| Sample: |
Tyrosine--tRNA ligase, cytoplasmic dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
|
|
|
|
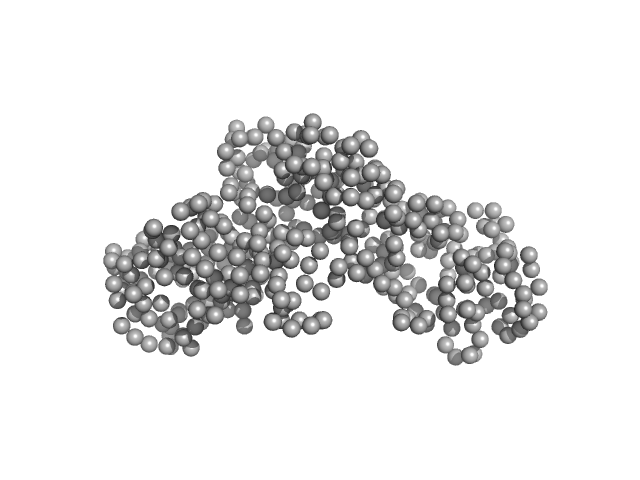
|
| Sample: |
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
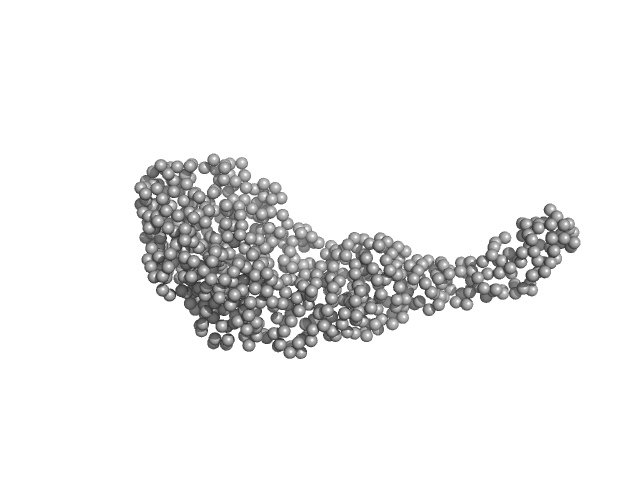
|
| Sample: |
Tegument protein UL21 (C-terminal domain) monomer, 29 kDa Human herpesvirus 1 … protein
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
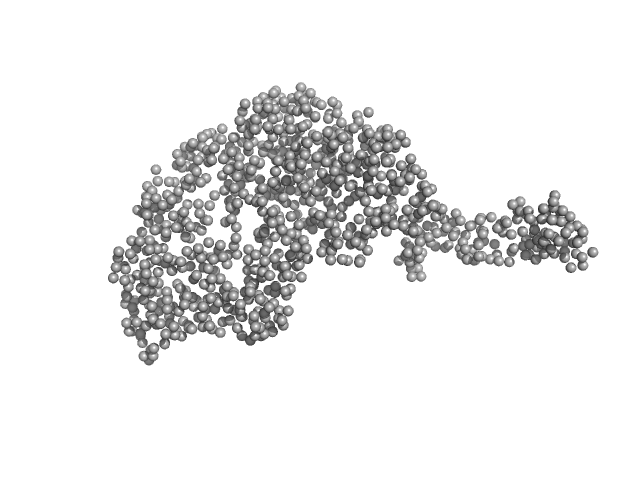
|
| Sample: |
Tegument protein UL21 monomer, 59 kDa Human alphaherpesvirus 1 … protein
Ceramide transfer protein (recombinant CERTL: amino acids 20-130 and 351-624) monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.5, 500 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Sep 25
|
Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT
Journal of Biological Chemistry :102589 (2022)
Benedyk T, Connor V, Caroe E, Shamin M, Svergun D, Deane J, Jeffries C, Crump C, Graham S
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Potassium voltage-gated channel subfamily B member 1 pentamer, 73 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% v/v glycerol, 0.01% Triton X-100, 10 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Jul 13
|
Pentameric assembly of the Kv2.1 tetramerization domain
Acta Crystallographica Section D Structural Biology 78(6):792-802 (2022)
Xu Z, Khan S, Schnicker N, Baker S
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nocturnin monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 10% glycerol, 5 mM MgCl2, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Feb 7
|
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.
Biochemistry (2022)
Wickramaratne AC, Li L, Hopkins JB, Joachimiak LA, Green CB
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nocturnin monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 10% glycerol, 5 mM MgCl2, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Feb 7
|
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.
Biochemistry (2022)
Wickramaratne AC, Li L, Hopkins JB, Joachimiak LA, Green CB
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nocturnin - N terminus truncated (118-NOCT) monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM KCl, 10% glycerol, 5 mM MgCl2, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Feb 7
|
The Disordered Amino Terminus of the Circadian Enzyme Nocturnin Modulates Its NADP(H) Phosphatase Activity by Changing Protein Dynamics.
Biochemistry (2022)
Wickramaratne AC, Li L, Hopkins JB, Joachimiak LA, Green CB
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1 (H123R) dimer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Oct 8
|
Structural insights into Charcot-Marie-Tooth disease-linked mutations in human GDAP1.
FEBS Open Bio (2022)
Sutinen A, Nguyen GTT, Raasakka A, Muruganandam G, Loris R, Ylikallio E, Tyynismaa H, Bartesaghi L, Ruskamo S, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1 (R120W) dimer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 1
|
Structural insights into Charcot-Marie-Tooth disease-linked mutations in human GDAP1.
FEBS Open Bio (2022)
Sutinen A, Nguyen GTT, Raasakka A, Muruganandam G, Loris R, Ylikallio E, Tyynismaa H, Bartesaghi L, Ruskamo S, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
105 |
nm3 |
|
|