|
|
|
|
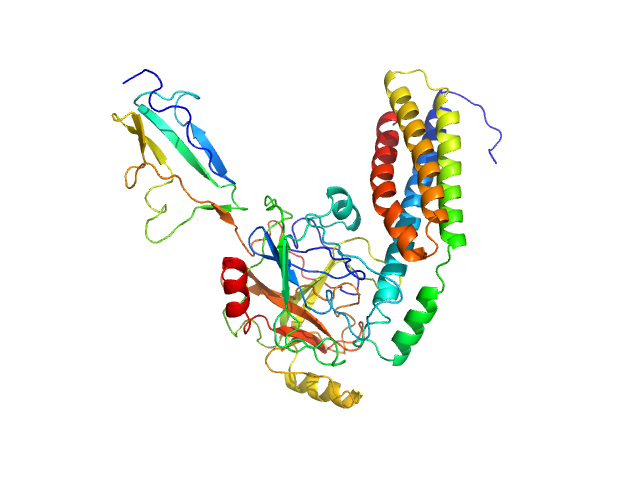
|
| Sample: |
Fibronectin-binding protein BBK32 monomer, 17 kDa Borrelia burgdorferi (strain … protein
Complement C1r subcomponent monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 140 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Sep 24
|
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
J Immunol 207(11):2856-2867 (2021)
Garrigues RJ, Powell-Pierce AD, Hammel M, Skare JT, Garcia BL
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein tau, Tau35 fragment monomer, 27 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 8
|
The Disease Associated Tau35 Fragment has an Increased Propensity to Aggregate Compared to Full-Length Tau
Frontiers in Molecular Biosciences 8 (2021)
Lyu C, Da Vela S, Al-Hilaly Y, Marshall K, Thorogate R, Svergun D, Serpell L, Pastore A, Hanger D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein tau, 2N3R isoform monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 8
|
The Disease Associated Tau35 Fragment has an Increased Propensity to Aggregate Compared to Full-Length Tau
Frontiers in Molecular Biosciences 8 (2021)
Lyu C, Da Vela S, Al-Hilaly Y, Marshall K, Thorogate R, Svergun D, Serpell L, Pastore A, Hanger D
|
| RgGuinier |
6.3 |
nm |
| Dmax |
19.8 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein tau, 2N4R isoform monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 8
|
The Disease Associated Tau35 Fragment has an Increased Propensity to Aggregate Compared to Full-Length Tau
Frontiers in Molecular Biosciences 8 (2021)
Lyu C, Da Vela S, Al-Hilaly Y, Marshall K, Thorogate R, Svergun D, Serpell L, Pastore A, Hanger D
|
| RgGuinier |
6.7 |
nm |
| Dmax |
23.5 |
nm |
| VolumePorod |
254 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Relaxin receptor 1 monomer, 8 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 10 mM CaCl2, 0.1%NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 6
|
Structural Insights into the Unique Modes of Relaxin-Binding and Tethered-Agonist Mediated Activation of RXFP1 and RXFP2.
J Mol Biol 433(21):167217 (2021)
Sethi A, Bruell S, Ryan T, Yan F, Tanipour MH, Mok YF, Draper-Joyce C, Khandokar Y, Metcalfe RD, Griffin MDW, Scott DJ, Hossain MA, Petrie EJ, Bathgate RAD, Gooley PR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Relaxin receptor 2 monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 10 mM CaCl2, 0.1%NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 6
|
Structural Insights into the Unique Modes of Relaxin-Binding and Tethered-Agonist Mediated Activation of RXFP1 and RXFP2.
J Mol Biol 433(21):167217 (2021)
Sethi A, Bruell S, Ryan T, Yan F, Tanipour MH, Mok YF, Draper-Joyce C, Khandokar Y, Metcalfe RD, Griffin MDW, Scott DJ, Hossain MA, Petrie EJ, Bathgate RAD, Gooley PR
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
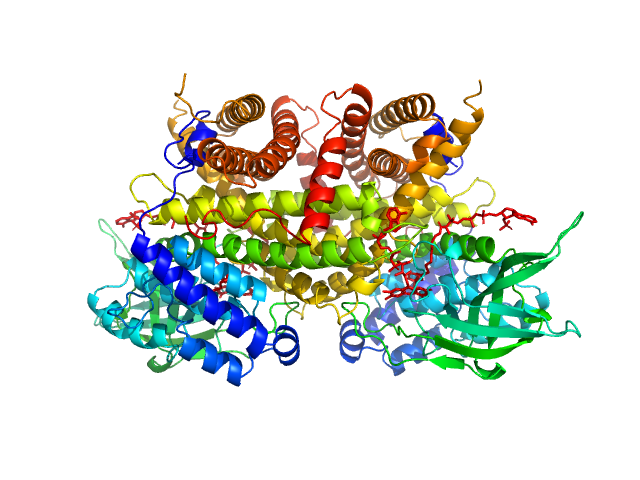
|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
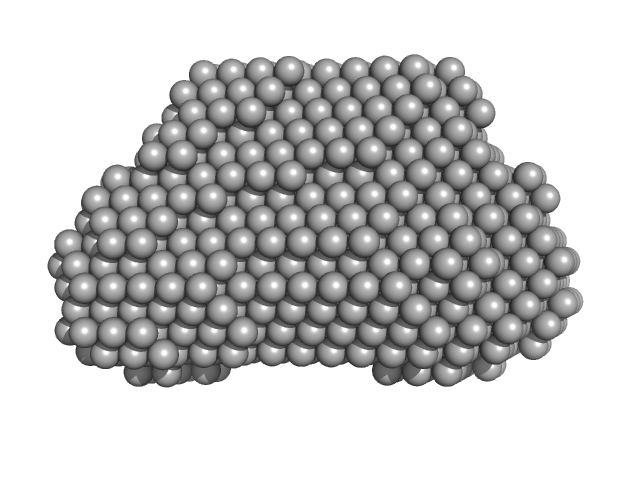
|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 130 kDa Homo sapiens protein
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial dimer, 93 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
514 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial tetramer, 259 kDa Homo sapiens protein
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial tetramer, 184 kDa Homo sapiens protein
Complex I intermediate-associated protein 30, mitochondrial tetramer, 190 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
6.6 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
1340 |
nm3 |
|
|
|
|
|
|
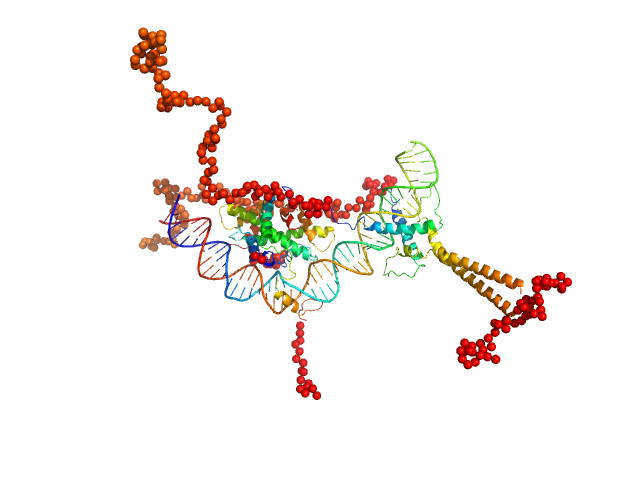
|
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
155 |
nm3 |
|
|