|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.9 |
nm |
|
|
|
|
|
|
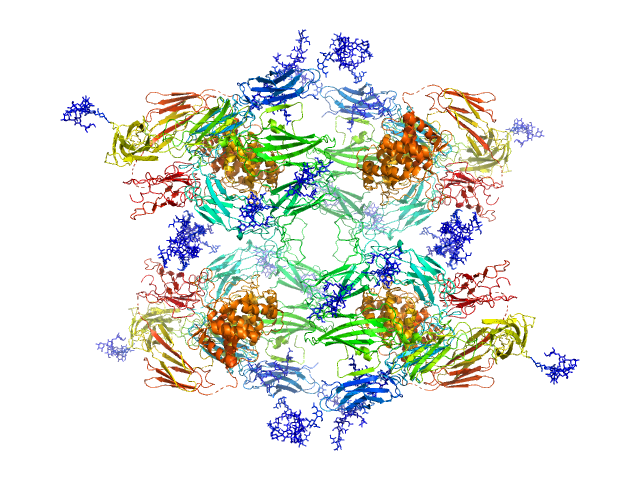
|
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
7.7 |
nm |
| Dmax |
22.7 |
nm |
|
|
|
|
|
|
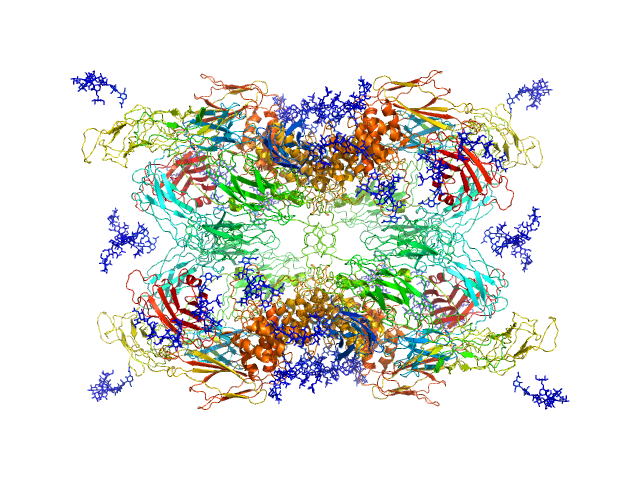
|
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.7 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
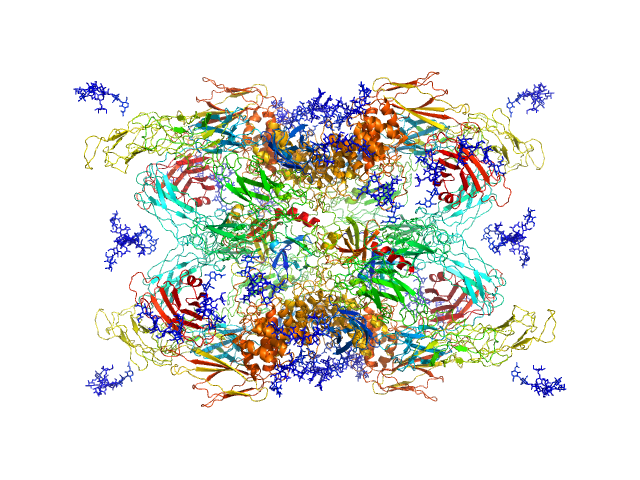
|
| Sample: |
Cationic trypsin dimer, 52 kDa Bos taurus protein
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 15
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
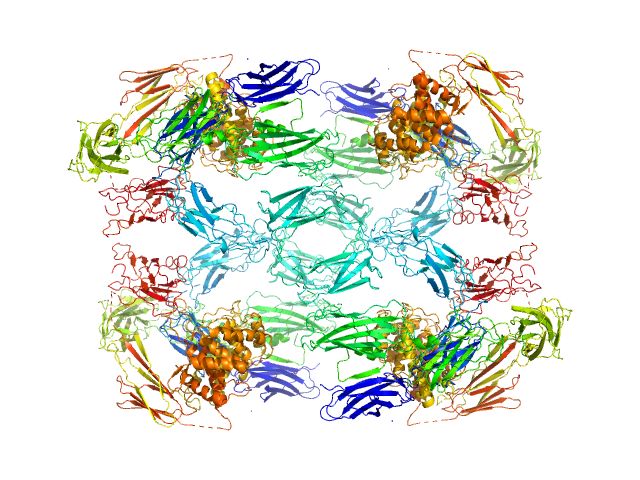
|
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2019 Apr 12
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
7.6 |
nm |
| Dmax |
20.2 |
nm |
|
|
|
|
|
|
|
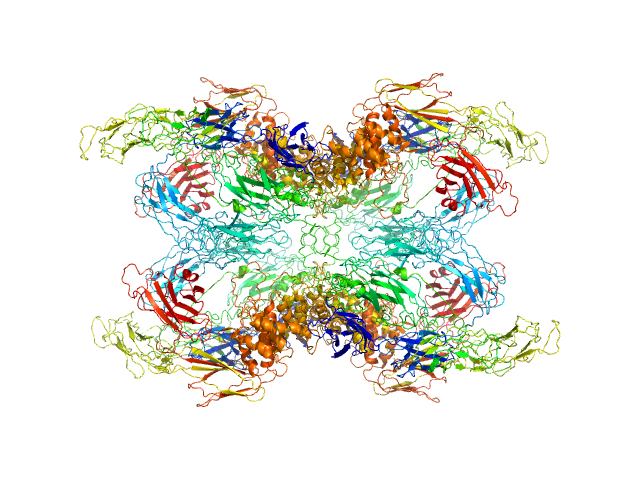
| Sample: |
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2019 Apr 13
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
19.7 |
nm |
|
|
|
|
|
|
|
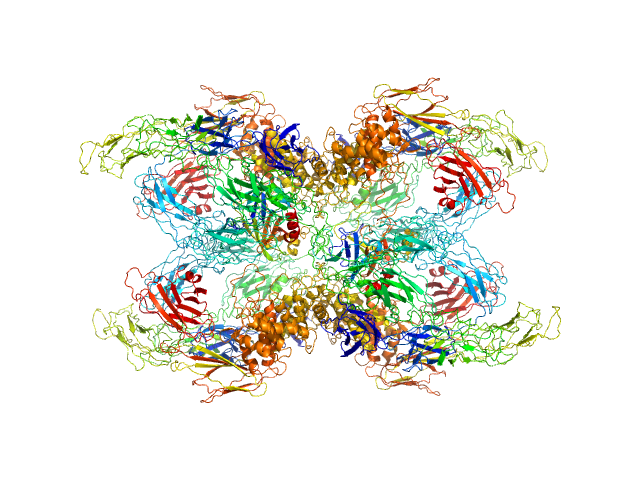
| Sample: |
Cationic trypsin dimer, 52 kDa Bos taurus protein
Alpha-2-macroglobulin tetramer, 643 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Bruker Nanostar w Excillum source, Department of Chemistry, iNANO building, Aarhus Uinversity on 2020 Jan 22
|
Structural Investigations of Human A2M Identify a Hollow Native Conformation That Underlies Its Distinctive Protease-Trapping Mechanism.
Mol Cell Proteomics 20:100090 (2021)
Harwood SL, Lyngsø J, Zarantonello A, Kjøge K, Nielsen PK, Andersen GR, Pedersen JS, Enghild JJ
|
| RgGuinier |
6.6 |
nm |
| Dmax |
20.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
N-ter construct of FATZ-1 (alias myozenin-1 or calsarcin-2) monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 29
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
46 |
nm3 |
|
|