|
|
|
|
|
| Sample: |
Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 18
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
3.9 |
nm |
| Dmax |
17.3 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rod domain of α-actinin-2 dimer, 112 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 5
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
6.7 |
nm |
| Dmax |
27.2 |
nm |
| VolumePorod |
214 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Half dimer of α-actinin-2 monomer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 18
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
5.3 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
172 |
nm3 |
|
|
|
|
|
|
|
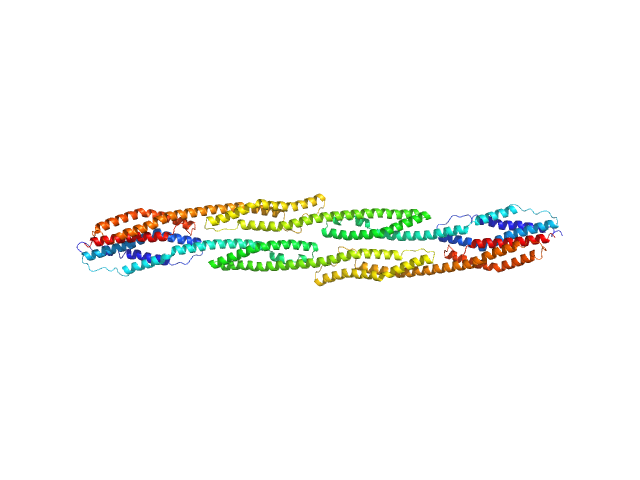
| Sample: |
Rod domain of α-actinin-2 dimer, 112 kDa Homo sapiens protein
Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2) dimer, 43 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 18
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
7.6 |
nm |
| Dmax |
28.4 |
nm |
| VolumePorod |
371 |
nm3 |
|
|
|
|
|
|
|
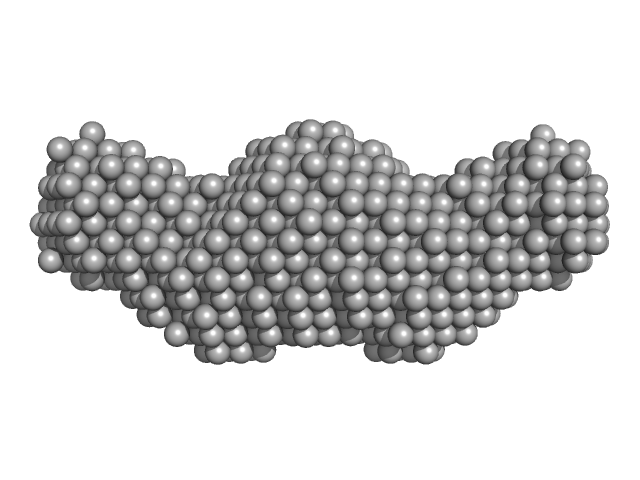
| Sample: |
Δ91 construct of FATZ-1 (alias myozenin-1 or calsarcin-2) monomer, 22 kDa Homo sapiens protein
Half dimer of α-actinin-2 monomer, 107 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 18
|
Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin.
Sci Adv 7(22) (2021)
Sponga A, Arolas JL, Schwarz TC, Jeffries CM, Rodriguez Chamorro A, Kostan J, Ghisleni A, Drepper F, Polyansky A, De Almeida Ribeiro E, Pedron M, Zawadzka-Kazimierczuk A, Mlynek G, Peterbauer T, Doto P, Schreiner C, Hollerl E, Mateos B, Geist L, Faulkner G, Kozminski W, Svergun DI, Warscheid B, Zagrovic B, Gautel M, Konrat R, Djinović-Carugo K
|
| RgGuinier |
5.8 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
242 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Zinc transporter 1 dimer, 37 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris, 150mM NaCl, 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 14
|
Heterologous Expression and Biochemical Characterization of the Human Zinc Transporter 1 (ZnT1) and Its Soluble C-Terminal Domain
Frontiers in Chemistry 9 (2021)
Cotrim C, Jarrott R, Whitten A, Choudhury H, Drew D, Martin J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polymerase delta-interacting protein 2 monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES pH 7.5, 500 mM NaCl, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, University of Huddersfield on 2019 Jul 18
|
Crystal structure and molecular dynamics of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability.
Protein Sci (2021)
Kulik AA, Maruszczak KK, Thomas DC, Nabi-Aldridge NLA, Carr M, Bingham RJ, Cooper CDO
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Prosaposin (Saposin A) monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 18
|
Stable Picodisc Assemblies from Saposin Proteins and Branched Detergents.
Biochemistry 60(14):1108-1119 (2021)
Kurgan KW, Chen B, Brown KA, Falco Cobra P, Ye X, Ge Y, Gellman SH
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Prosaposin (Saposin-A) dimer, 19 kDa Homo sapiens protein
1,2,5,6-tetra-β-D-glucopyranoside-3,4-O-Di-dodecyl-d-mannitol decamer, 12 kDa
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 18
|
Stable Picodisc Assemblies from Saposin Proteins and Branched Detergents.
Biochemistry 60(14):1108-1119 (2021)
Kurgan KW, Chen B, Brown KA, Falco Cobra P, Ye X, Ge Y, Gellman SH
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Prosaposin (Saposin-A) dimer, 19 kDa Homo sapiens protein
1,2,5,6-tetra-β-D-glucopyranoside-3,4-O-Di-tridecyl-d-mannitol decamer, 12 kDa
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Feb 18
|
Stable Picodisc Assemblies from Saposin Proteins and Branched Detergents.
Biochemistry 60(14):1108-1119 (2021)
Kurgan KW, Chen B, Brown KA, Falco Cobra P, Ye X, Ge Y, Gellman SH
|
| RgGuinier |
2.5 |
nm |
| Dmax |
6.7 |
nm |
|
|