|
|
|
|
|
| Sample: |
Histatin-3, His3-(20-43)-peptide monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 13
|
Comment on the Optimal Parameters to Derive Intrinsically Disordered Protein Conformational Ensembles from Small-Angle X-ray Scattering Data Using the Ensemble Optimization Method
Journal of Chemical Theory and Computation (2021)
Sagar A, Jeffries C, Petoukhov M, Svergun D, Bernadó P
|
| RgGuinier |
1.4 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histatin-3, His3-(20-43)-peptide monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 13
|
Comment on the Optimal Parameters to Derive Intrinsically Disordered Protein Conformational Ensembles from Small-Angle X-ray Scattering Data Using the Ensemble Optimization Method
Journal of Chemical Theory and Computation (2021)
Sagar A, Jeffries C, Petoukhov M, Svergun D, Bernadó P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histatin-3, His3-(20-43)-peptide monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Mar 13
|
Comment on the Optimal Parameters to Derive Intrinsically Disordered Protein Conformational Ensembles from Small-Angle X-ray Scattering Data Using the Ensemble Optimization Method
Journal of Chemical Theory and Computation (2021)
Sagar A, Jeffries C, Petoukhov M, Svergun D, Bernadó P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|
|
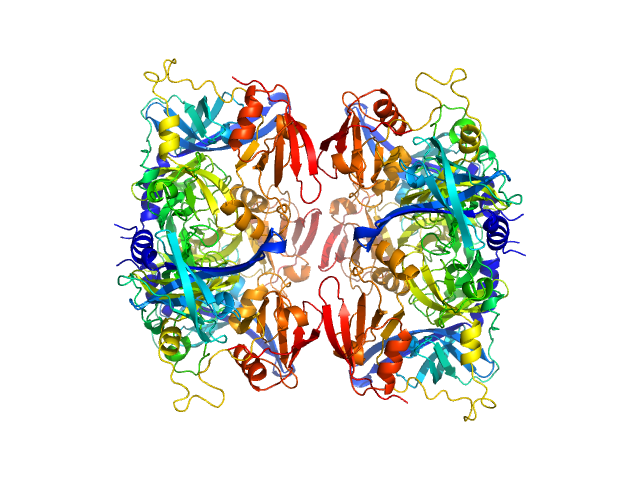
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
|
|
|
|
|
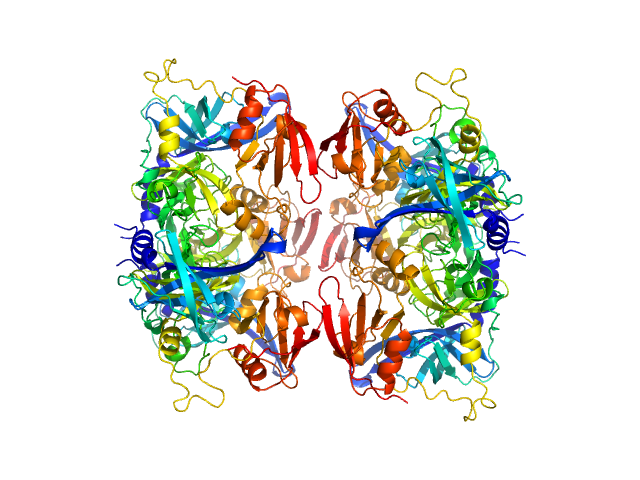
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
|
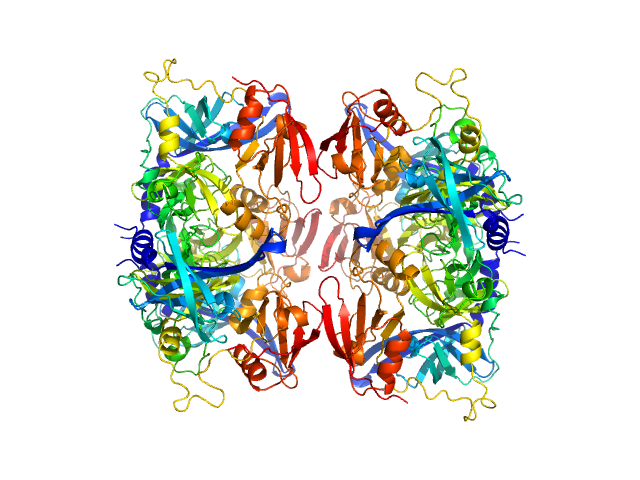
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
203 |
nm3 |
|
|
|
|
|
|
|
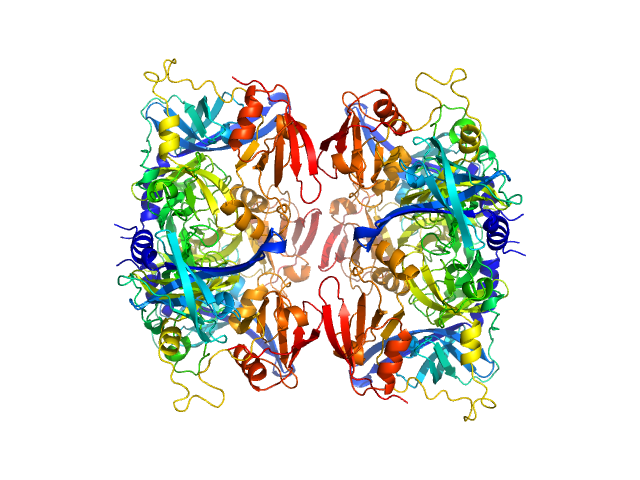
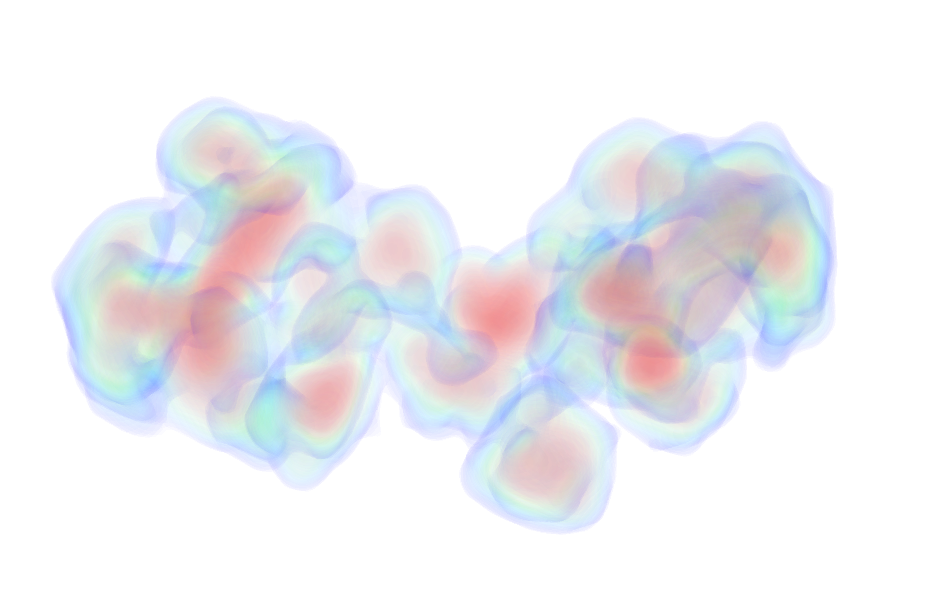
| Sample: |
Serine protease HTRA2, mitochondrial hexamer, 210 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES-NaOH (pH 7.4), 120 mM NaCl, 1 mM EDTA, and 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 1
|
Oligomeric assembly regulating mitochondrial HtrA2 function as examined by methyl-TROSY NMR
Proceedings of the National Academy of Sciences 118(11):e2025022118 (2021)
Toyama Y, Harkness R, Lee T, Maynes J, Kay L
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
|
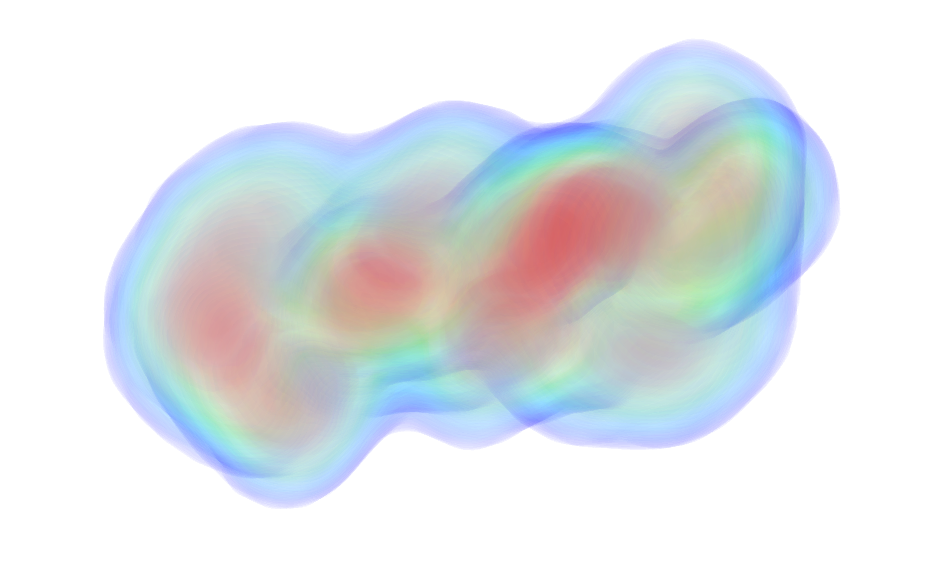
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 4 mM CaCl₂, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Sep 24
|
A High‐Affinity Calmodulin‐Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
Advanced Science :2003630 (2021)
Voegele A, Sadi M, O'Brien D, Gehan P, Raoux‐Barbot D, Davi M, Hoos S, Brûlé S, Raynal B, Weber P, Mechaly A, Haouz A, Rodriguez N, Vachette P, Durand D, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calmodulin-1 monomer, 17 kDa Homo sapiens protein
Bifunctional hemolysin/adenylate cyclase monomer, 3 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 4 mM CaCl₂, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Sep 24
|
A High‐Affinity Calmodulin‐Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells
Advanced Science :2003630 (2021)
Voegele A, Sadi M, O'Brien D, Gehan P, Raoux‐Barbot D, Davi M, Hoos S, Brûlé S, Raynal B, Weber P, Mechaly A, Haouz A, Rodriguez N, Vachette P, Durand D, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
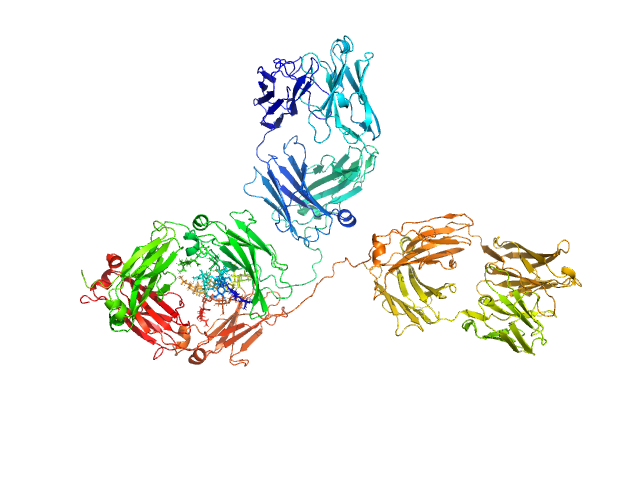
|
| Sample: |
Immunoglobulin G subclass 1, 148 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, and 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 20
|
Solution structure of deglycosylated human IgG1 shows the role of CH2 glycans in its conformation
Biophysical Journal (2021)
Spiteri V, Doutch J, Rambo R, Gor J, Dalby P, Perkins S
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
269 |
nm3 |
|
|