|
|
|
|
|
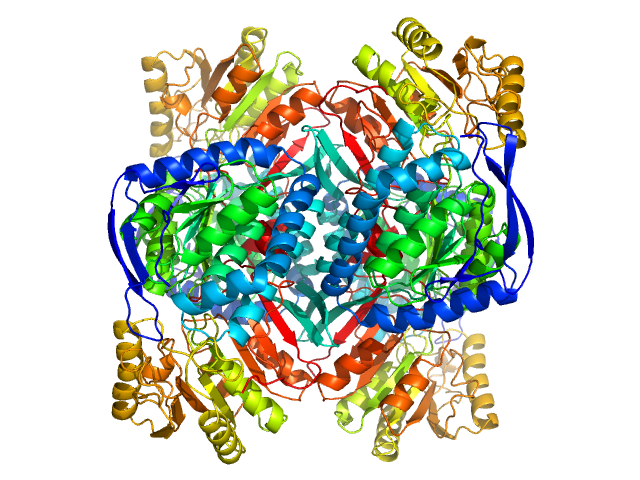
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
|
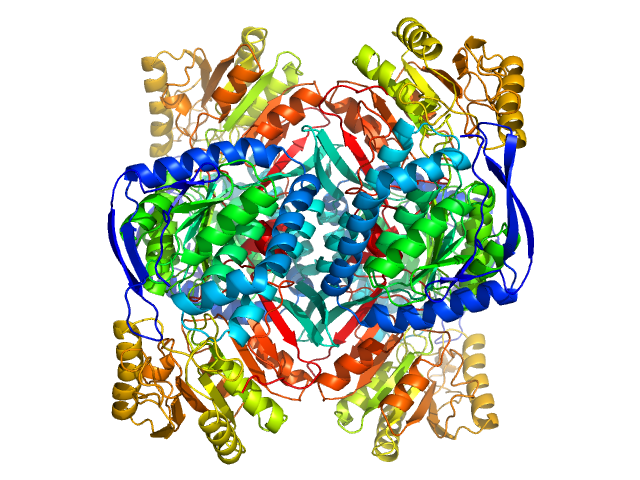
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
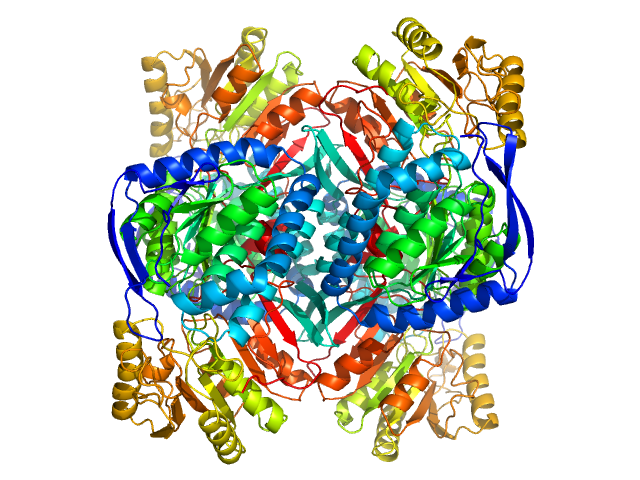
| Sample: |
4-trimethylaminobutyraldehyde dehydrogenase tetramer, 215 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 600 mM NaCl, 2% (v/v) glycerol, 1 mM DTT, 1 mM NAD+, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 28
|
Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch Biochem Biophys :108477 (2020)
Wyatt JW, Korasick DA, Qureshi IA, Campbell AC, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagen alpha-3(VI) chain, N2 domain monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM TRIS, pH 7.4, 150mM NaCl 3% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 4
|
Structure of a collagen VI α3 chain VWA domain array: adaptability and functional implications of myopathy causing mutations
Journal of Biological Chemistry :jbc.RA120.014865 (2020)
Solomon-Degefa H, Gebauer J, Jeffries C, Freiburg C, Meckelburg P, Bird L, Baumann U, Svergun D, Owens R, Werner J, Behrmann E, Paulsson M, Wagener R
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
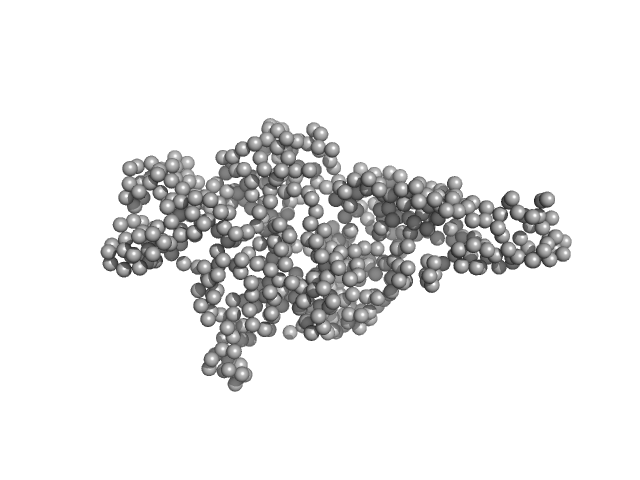
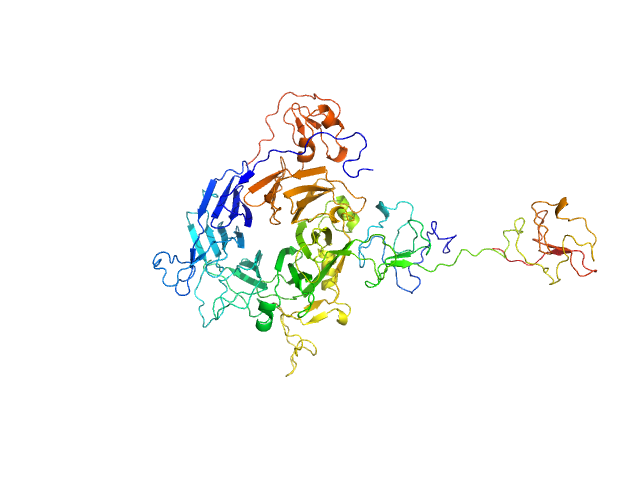
| Sample: |
Hepatocyte growth factor receptor monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 27
|
Dimerization of kringle 1 domain from hepatocyte growth factor/scatter factor provides a potent MET receptor agonist
Life Science Alliance 5(12):e202201424 (2022)
de Nola G, Leclercq B, Mougel A, Taront S, Simonneau C, Forneris F, Adriaenssens E, Drobecq H, Iamele L, Dubuquoy L, Melnyk O, Gherardi E, de Jonge H, Vicogne J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Minimal hepatocyte growth factor mimic K1K1 monomer, 19 kDa synthetic construct protein
Hepatocyte growth factor receptor monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jan 23
|
Dimerization of kringle 1 domain from hepatocyte growth factor/scatter factor provides a potent MET receptor agonist
Life Science Alliance 5(12):e202201424 (2022)
de Nola G, Leclercq B, Mougel A, Taront S, Simonneau C, Forneris F, Adriaenssens E, Drobecq H, Iamele L, Dubuquoy L, Melnyk O, Gherardi E, de Jonge H, Vicogne J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Oct 19
|
The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity andcontributes to membrane binding.
J Biol Chem (2020)
Ravala SK, Hopkins JB, Plescia CB, Allgood SR, Kane MA, Cash JN, Stahelin RV, Tesmer JJG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 monomer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Oct 19
|
The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity andcontributes to membrane binding.
J Biol Chem (2020)
Ravala SK, Hopkins JB, Plescia CB, Allgood SR, Kane MA, Cash JN, Stahelin RV, Tesmer JJG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
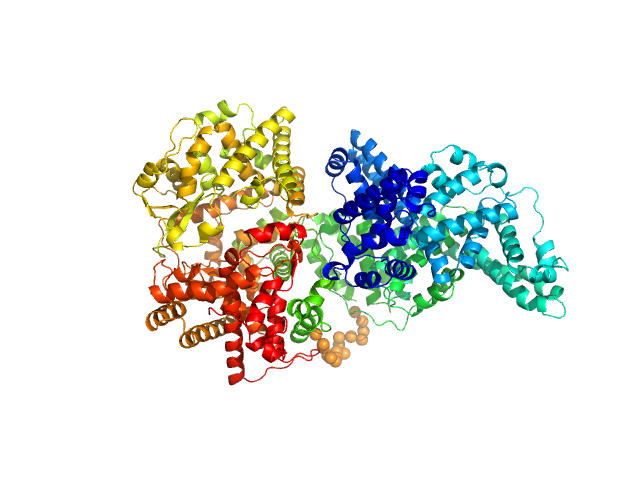
| Sample: |
Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Oct 19
|
The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity andcontributes to membrane binding.
J Biol Chem (2020)
Ravala SK, Hopkins JB, Plescia CB, Allgood SR, Kane MA, Cash JN, Stahelin RV, Tesmer JJG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM histidine, pH: 5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 21
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
267 |
nm3 |
|
|