|
|
|
|
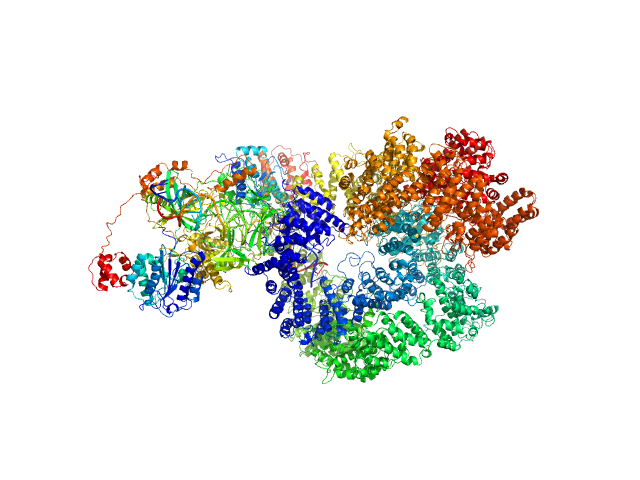
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
6.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
1090 |
nm3 |
|
|
|
|
|
|
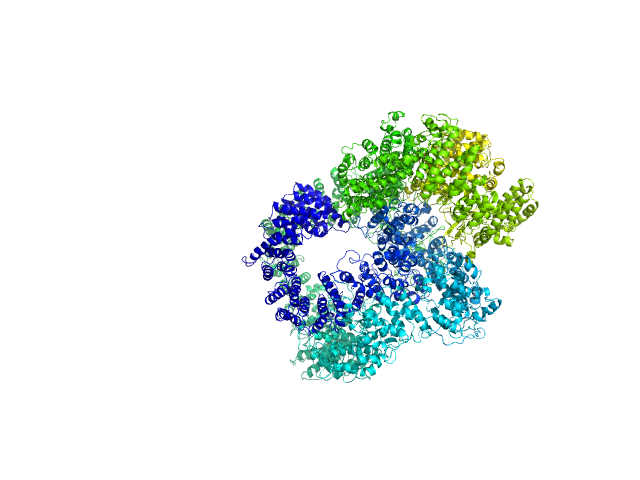
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
7.5 |
nm |
| Dmax |
29.4 |
nm |
| VolumePorod |
1440 |
nm3 |
|
|
|
|
|
|
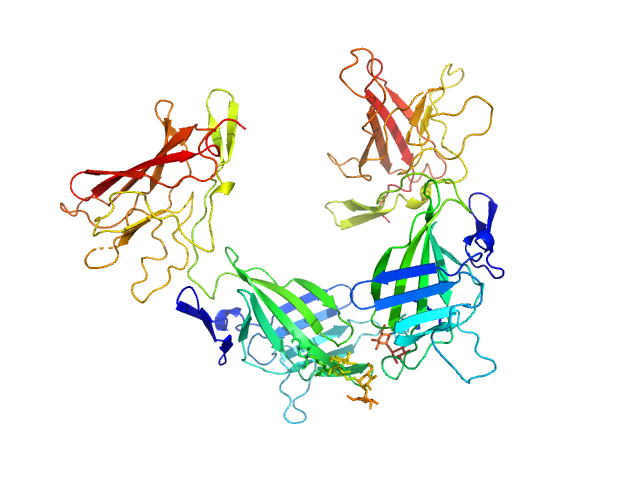
|
| Sample: |
Cation-independent mannose-6-phosphate receptor dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
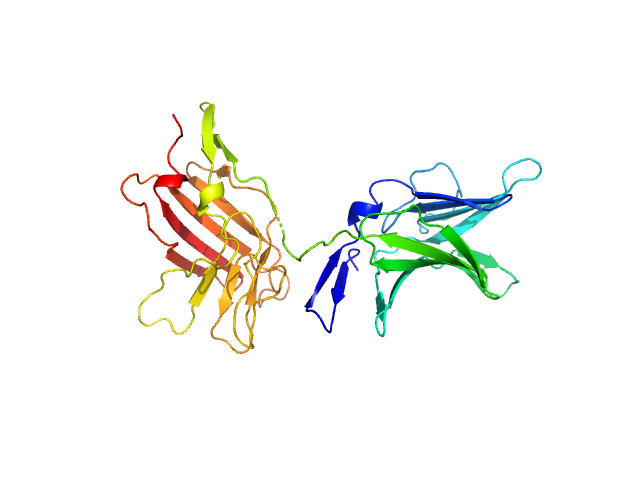
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
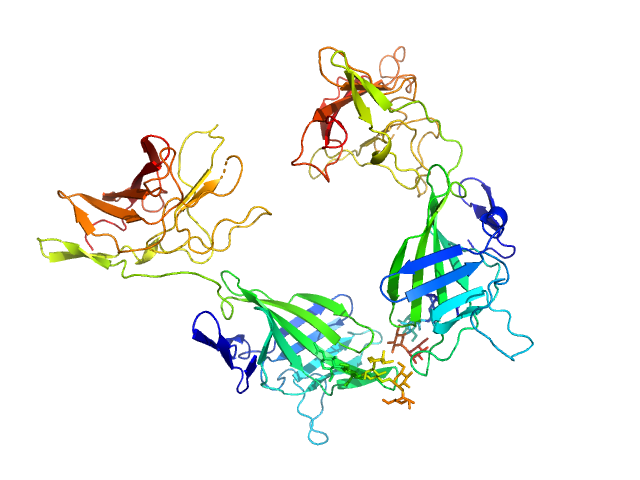
|
| Sample: |
Cation-independent mannose-6-phosphate receptor dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
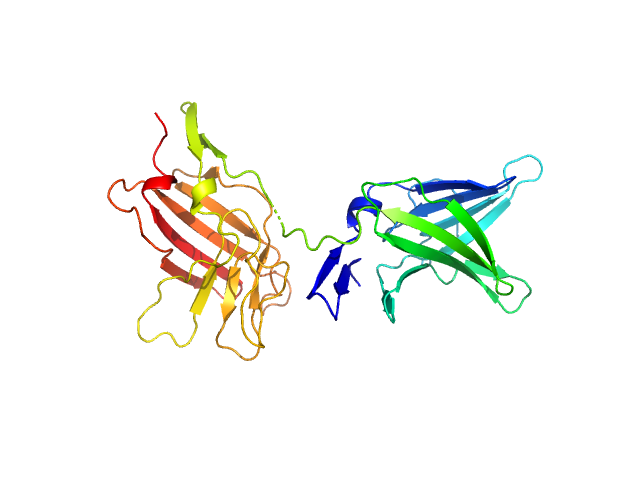
|
| Sample: |
Cation-independent mannose-6-phosphate receptor monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7–11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9
Structure (2020)
Bochel A, Williams C, McCoy A, Hoppe H, Winter A, Nicholls R, Harlos K, Jones E, Berger I, Hassan A, Crump M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Vitamin K-dependent protein C monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 145 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 14
|
Zymogen and activated protein C have similar structural architecture.
J Biol Chem (2020)
Stojanovski BM, Pelc LA, Zuo X, Di Cera E
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|
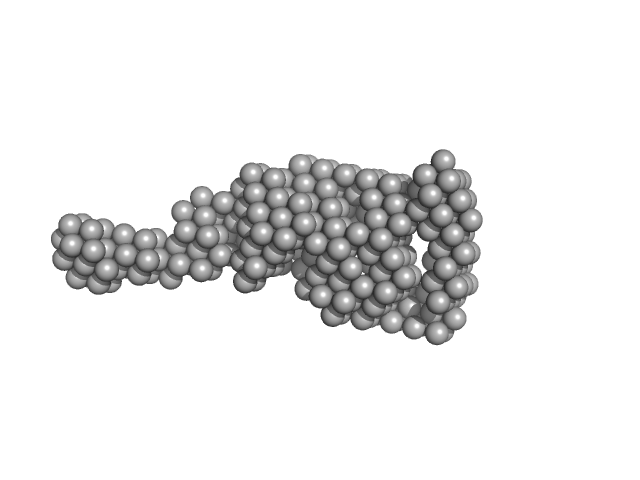
|
| Sample: |
Vitamin K-dependent protein C monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 145 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Aug 17
|
Zymogen and activated protein C have similar structural architecture.
J Biol Chem (2020)
Stojanovski BM, Pelc LA, Zuo X, Di Cera E
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|
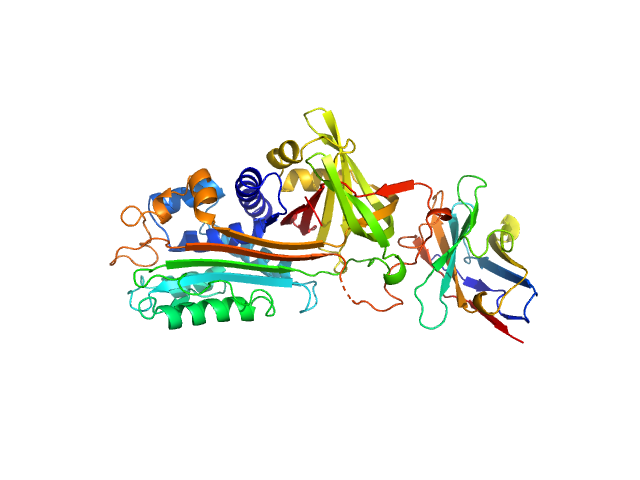
|
| Sample: |
Plasminogen activator inhibitor 1 monomer, 43 kDa Homo sapiens protein
VHH-s-a93 (Ig module Nb93) monomer, 13 kDa Vicugna pacos protein
|
| Buffer: |
30 mM BIS-TRIS pH 5.5, 300 mM sodium chloride, 5% v/v glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 5
|
Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity
International Journal of Molecular Sciences 21(16):5859 (2020)
Sillen M, Weeks S, Strelkov S, Declerck P
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Carbonic anhydrase 9 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Nov 16
|
Biophysical Characterization of Cancer-Related Carbonic Anhydrase IX.
Int J Mol Sci 21(15) (2020)
Koruza K, Murray AB, Mahon BP, Hopkins JB, Knecht W, McKenna R, Fisher SZ
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
71 |
nm3 |
|
|