|
|
|
|
|
| Sample: |
Protein jagged-1 cysteine-rich domain, 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 11
|
Notch-Jagged signaling complex defined by an interaction mosaic.
Proc Natl Acad Sci U S A 118(30) (2021)
Zeronian MR, Klykov O, Portell I de Montserrat J, Konijnenberg MJ, Gaur A, Scheltema RA, Janssen BJC
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein jagged-1 cysteine-rich domain, 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 11
|
Notch-Jagged signaling complex defined by an interaction mosaic.
Proc Natl Acad Sci U S A 118(30) (2021)
Zeronian MR, Klykov O, Portell I de Montserrat J, Konijnenberg MJ, Gaur A, Scheltema RA, Janssen BJC
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein jagged-1 cysteine-rich domain, 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Dec 11
|
Notch-Jagged signaling complex defined by an interaction mosaic.
Proc Natl Acad Sci U S A 118(30) (2021)
Zeronian MR, Klykov O, Portell I de Montserrat J, Konijnenberg MJ, Gaur A, Scheltema RA, Janssen BJC
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
|
| Buffer: |
10mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Characterization of murine prion protein
Stefano Da Vela
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Interferon-activable protein 204 monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 100 mM KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2013 Mar 14
|
Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res (2021)
Fan X, Jiang J, Zhao D, Chen F, Ma H, Smith P, Unterholzner L, Xiao TS, Jin T
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Homo sapiens protein
HUI-018 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2013 Mar 7
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Homo sapiens protein
HUI-018 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at I911-4, MAX IV on 2013 Mar 7
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
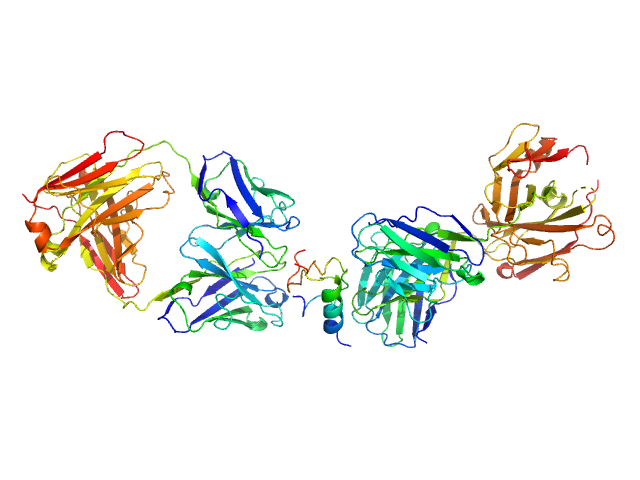
|
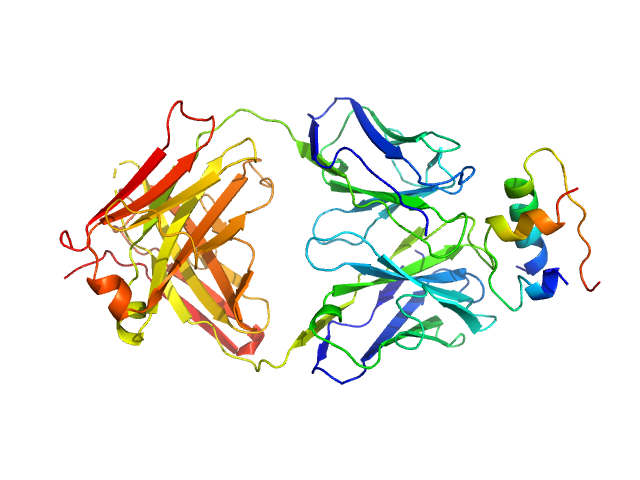
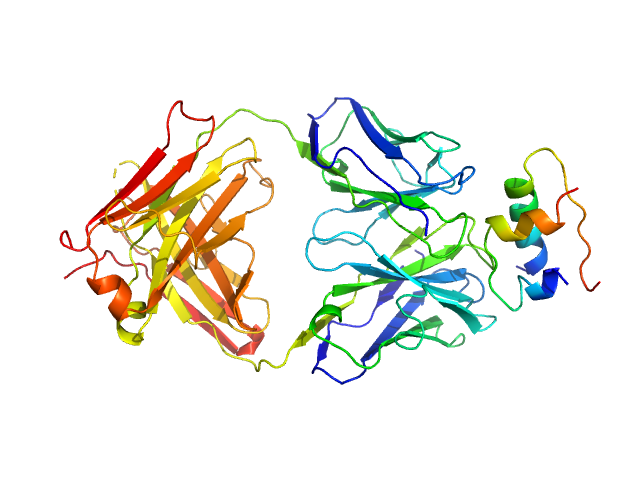
| Sample: |
HUI-018 Fab monomer, 47 kDa Mus musculus protein
Insulin (Insulin B chain and Insulin A chain) monomer, 6 kDa Sus scrofa protein
OXI-005 Fab monomer, 47 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Novo Nordisk A/S on 2019 Mar 28
|
Insulin Binding to the Analytical Antibody Sandwich Pair OXI
‐005 and HUI
‐018 – Epitope Mapping and Binding Properties
Protein Science (2020)
Johansson E, Wu X, Yu B, Yang Z, Cao Z, Wiberg C, Jeppesen C, Poulsen F
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
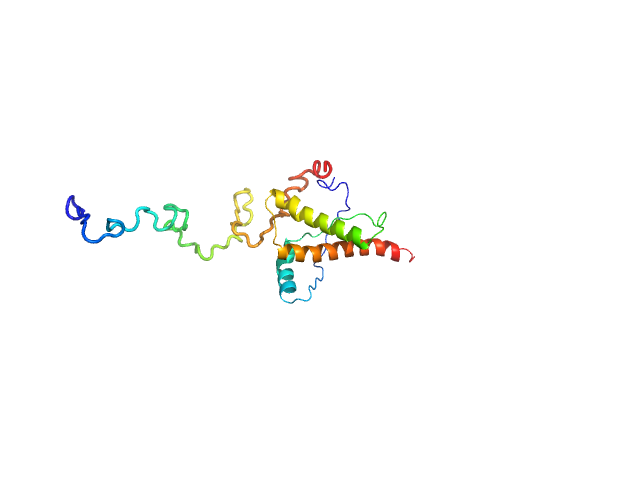
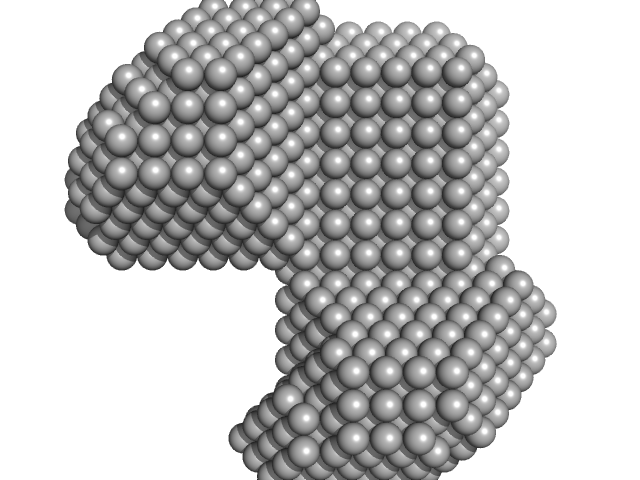
| Sample: |
LIM/homeobox protein Lhx3 monomer, 10 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
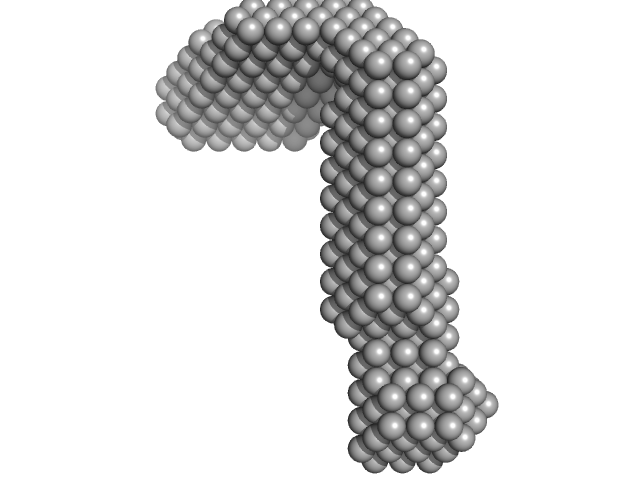
|
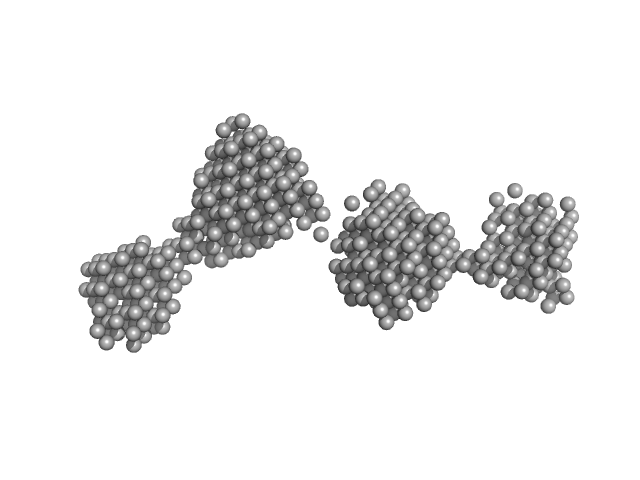
| Sample: |
LIM/homeobox protein Lhx3 monomer, 23 kDa Mus musculus protein
Insulin gene enhancer protein ISL-1 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM sodium phosphate monobasic/dibasic, 100 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Nov 2
|
Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification
Journal of Structural Biology: X :100043 (2020)
Smith N, Wilkinson-White L, Kwan A, Trewhella J, Matthews J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|