|
|
|
|

|
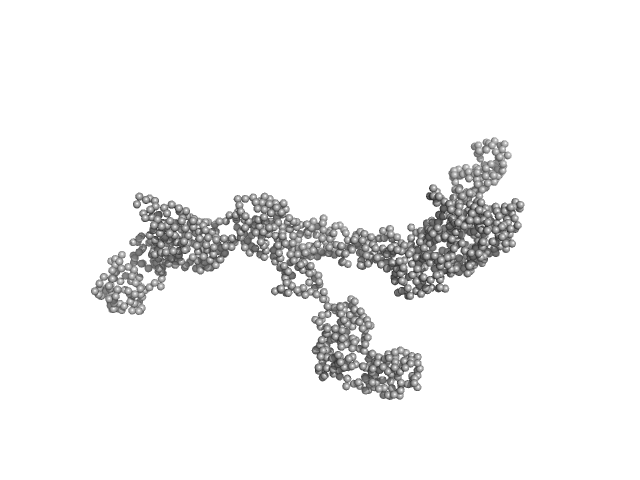
| Sample: |
Transcription intermediary factor 1-beta dimer, 82 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris 300 mM NaCl 0.1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Aug 10
|
A Dissection of Oligomerisation by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
J Mol Biol (2019)
Sun Y, Keown JR, Black MM, Raclot C, Demarais N, Trono D, Turelli P, Goldstone DC
|
| RgGuinier |
7.0 |
nm |
| Dmax |
23.2 |
nm |
| VolumePorod |
232 |
nm3 |
|
|
|
|
|
|
|
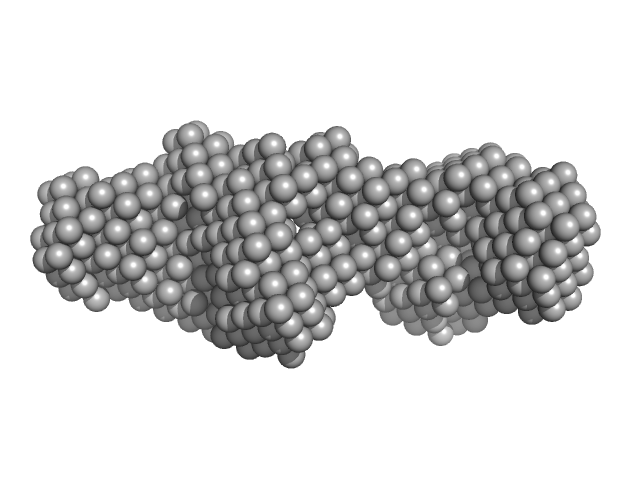
| Sample: |
Transcription intermediary factor 1-beta dimer, 82 kDa Mus musculus protein
Zinc finger protein 809 N-terminal MBP fusion monomer, 52 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris 300 mM NaCl 0.1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Aug 10
|
A Dissection of Oligomerisation by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.
J Mol Biol (2019)
Sun Y, Keown JR, Black MM, Raclot C, Demarais N, Trono D, Turelli P, Goldstone DC
|
| RgGuinier |
6.4 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
|
|
|
|
|
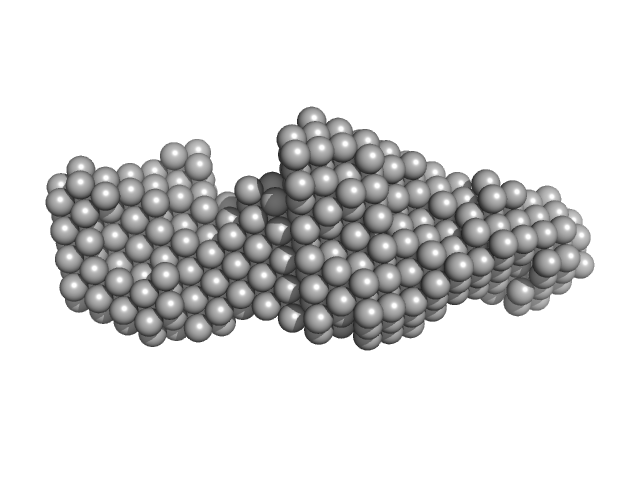
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
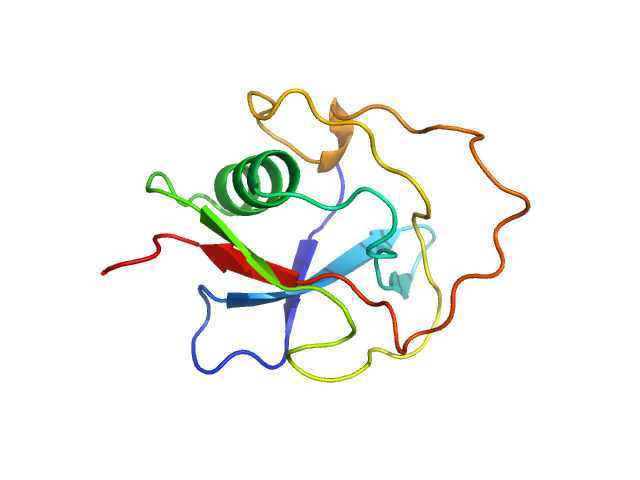
| Sample: |
LIM/homeobox protein Lhx4 monomer, 15 kDa Mus musculus protein
Insulin gene enhancer protein ISL-2 (R282G) monomer, 4 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins (2019)
Stokes PH, Robertson NO, Silva AP, Estephan T, Trewhella J, Guss JM, Matthews JM
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mouse Neurotrypsin Scavenger Receptor Cysteine-Rich Domain 3 monomer, 13 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES, 0.1 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 20
|
Structural characterization of the third scavenger receptor cysteine-rich domain of murine neurotrypsin.
Protein Sci 28(4):746-755 (2019)
Canciani A, Catucci G, Forneris F
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 4 dimer, 79 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 0.02% NaN3,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 13
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
183 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 4 dimer, 109 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris HCl, 100 mM NaCl, 0.02% NaN3,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 11
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
313 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Leucine-rich repeat and fibronectin type-III domain-containing protein 5 dimer, 82 kDa Mus musculus protein
|
| Buffer: |
30 mM Tris-Cl, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jun 8
|
The structure of SALM5 suggests a dimeric assembly for the presynaptic RPTP ligand recognition.
Protein Eng Des Sel (2018)
Karki S, Paudel P, Sele C, Shkumatov AV, Kajander T
|
| RgGuinier |
3.6 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein A-I dimer, 50 kDa Mus musculus protein
1,2-dimyristoyl-sn-glycero-3-phosphocholine, 92 kDa
|
| Buffer: |
PBS in D2O, pH: 7.4 |
| Experiment: |
SANS
data collected at NG7, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2015 Nov 25
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.1 |
nm |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein A-I dimer, 50 kDa Mus musculus protein
1,2-dimyristoyl-sn-glycero-3-phosphocholine, 92 kDa
|
| Buffer: |
PBS in D2O, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXSLab Bio-Ganesha, Institute for Bioscience and Biotechnology Research, National Institute of Standards and Technology (IBBR/NIST) on 2015 Nov 24
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
12.0 |
nm |
|
|