|
|
|
|
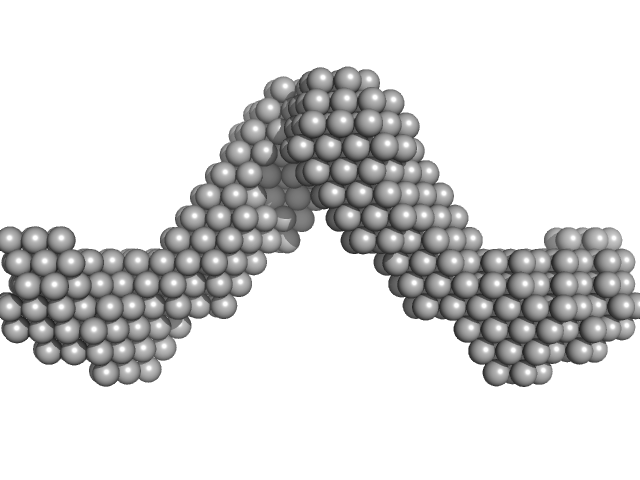
|
| Sample: |
Noelin tetramer, 256 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 6
|
Olfactomedin-1 Has a V-shaped Disulfide-linked Tetrameric Structure.
J Biol Chem 290(24):15092-101 (2015)
Pronker MF, Bos TG, Sharp TH, Thies-Weesie DM, Janssen BJ
|
| RgGuinier |
8.5 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
616 |
nm3 |
|
|
|
|
|
|
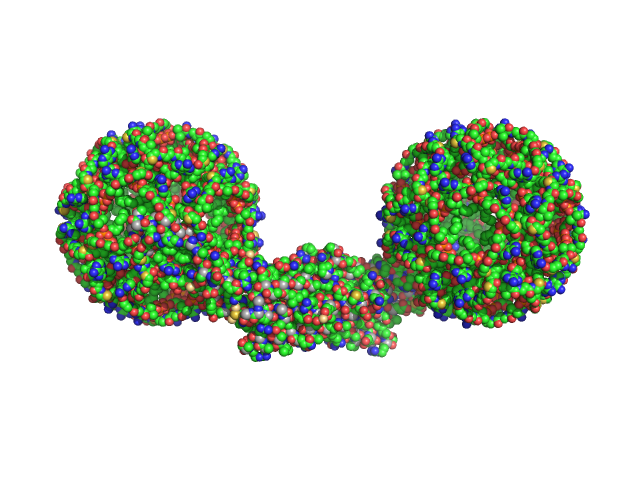
|
| Sample: |
Endophilin-A1 BAR domain dimer, 58 kDa Mus musculus protein
Arachidonyl-CoA, 60 kDa
|
| Buffer: |
50 mM TRIS-HCL 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 27
|
Endophilin-A1 BAR domain interaction with arachidonyl CoA.
Front Mol Biosci 1:20 (2014)
Petoukhov MV, Weissenhorn W, Svergun DI
|
| RgGuinier |
5.9 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
480 |
nm3 |
|
|
|
|
|
|
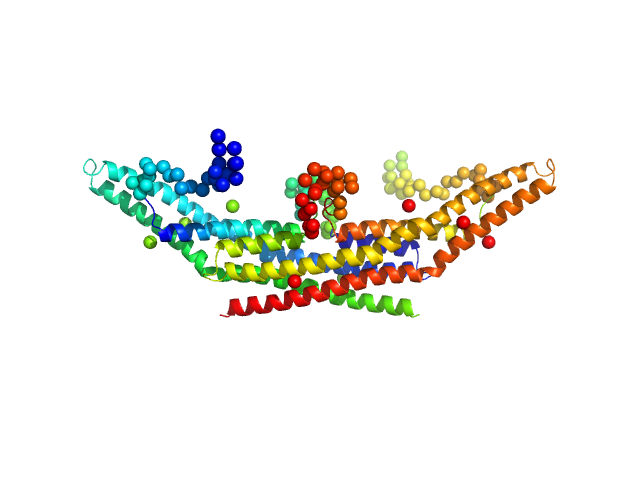
|
| Sample: |
Endophilin-A1 BAR domain dimer, 58 kDa Mus musculus protein
|
| Buffer: |
50 mM TRIS-HCL 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 13
|
Endophilin-A1 BAR domain interaction with arachidonyl CoA.
Front Mol Biosci 1:20 (2014)
Petoukhov MV, Weissenhorn W, Svergun DI
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
GTP-binding nuclear protein Ran monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
GTP-binding nuclear protein Ran monomer, 24 kDa Homo sapiens protein
Snurportin-1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
Snurportin-1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 10
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 7
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 8
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform 3 of Rap guanine nucleotide exchange factor 4 monomer, 114 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Utah on 2008 Aug 11
|
Mechanism of Epac activation: structural and functional analyses of Epac2 hinge mutants with constitutive and reduced activities.
J Biol Chem 284(35):23644-51 (2009)
Tsalkova T, Blumenthal DK, Mei FC, White MA, Cheng X
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
145 |
nm3 |
|
|