|
|
|
|

|
| Sample: |
AD11 Fab dimer, 50 kDa Mus musculus protein
|
| Buffer: |
50 mM Na-phosphate 1mM ethylenediaminetetraacetic aci, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 20
|
Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J Mol Biol 381(4):881-96 (2008)
Covaceuszach S, Cassetta A, Konarev PV, Gonfloni S, Rudolph R, Svergun DI, Lamba D, Cattaneo A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
NGF dimer, 50 kDa Mus musculus protein
|
| Buffer: |
10 mM Na-phosphate 150 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 20
|
Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J Mol Biol 381(4):881-96 (2008)
Covaceuszach S, Cassetta A, Konarev PV, Gonfloni S, Rudolph R, Svergun DI, Lamba D, Cattaneo A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
AD11 Fab dimer, 50 kDa Mus musculus protein
NGF dimer, 50 kDa Mus musculus protein
|
| Buffer: |
30 mM Na-phosphate 75 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Dec 20
|
Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J Mol Biol 381(4):881-96 (2008)
Covaceuszach S, Cassetta A, Konarev PV, Gonfloni S, Rudolph R, Svergun DI, Lamba D, Cattaneo A
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
|
|
|
|
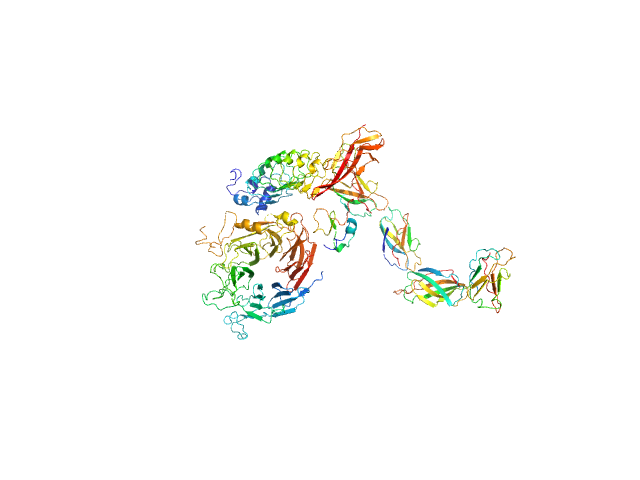
|
| Sample: |
Hepatocyte growth factor receptor monomer, 100 kDa Mus musculus protein
Internalin B monomer, 36 kDa Listeria monocytogenes serotype … protein
|
| Buffer: |
25mM Na-phosphate buffer, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Mar 1
|
X-ray and Neutron Small-Angle Scattering Analysis of the Complex Formed by the Met Receptor and the Listeria monocytogenes Invasion Protein InlB
Journal of Molecular Biology 377(2):489-500 (2008)
Niemann H, Petoukhov M, Härtlein M, Moulin M, Gherardi E, Timmins P, Heinz D, Svergun D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
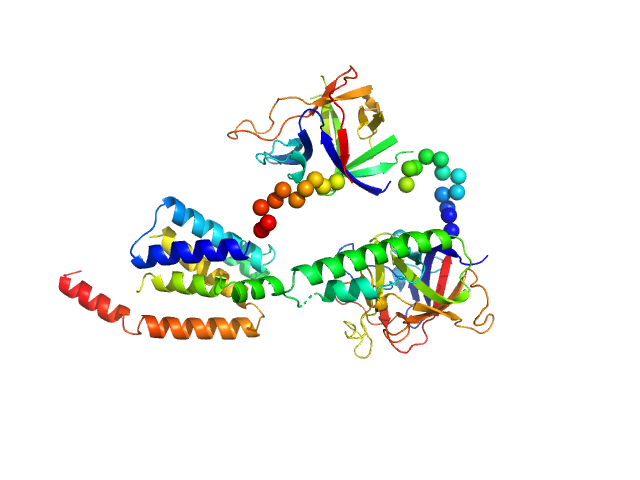
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 5 mM EGTA, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
|
|
|
|
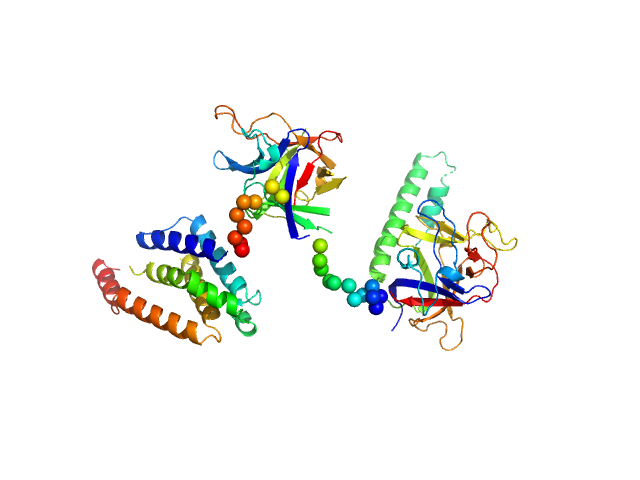
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 10 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
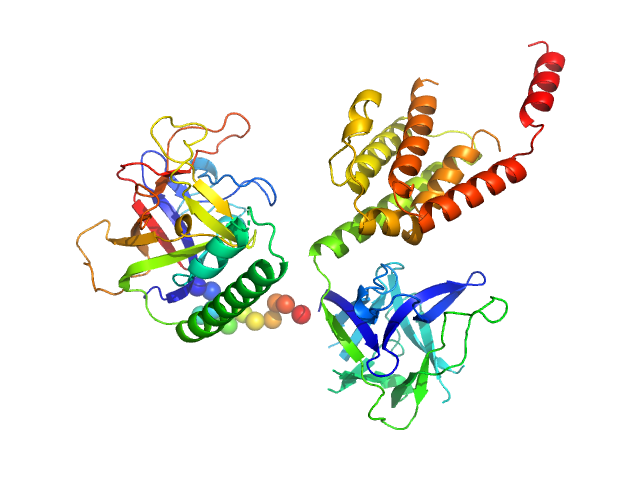
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 5 mM EGTA, 0.25 mM IP3, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
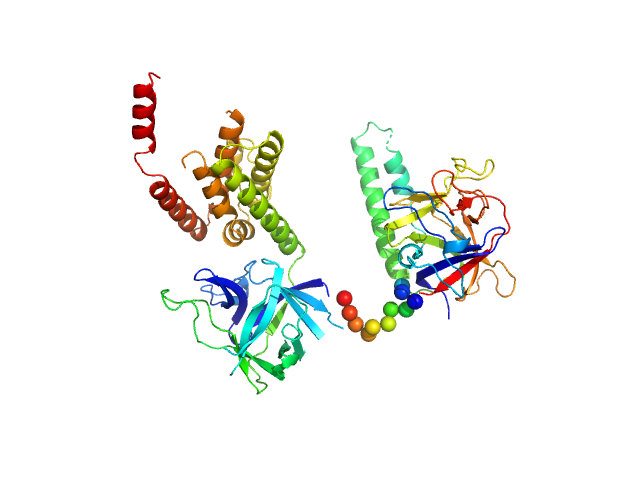
|
| Sample: |
N-terminal domains of the inositol 1,4,5-trisphosphate receptor type 1 monomer, 70 kDa Mus musculus protein
|
| Buffer: |
15 mM Tris, 300 mM NaCl, 1 mM TCEP, 10 mM CaCl2, 0.25 mM IP3, pH: 8 |
| Experiment: |
SAXS
data collected at Bruker Nanostar II, Australian Nuclear Science and Technology Organisation/Australian Centre for Neutron Scattering on 2006 Apr 12
|
Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor.
J Mol Biol 373(5):1269-80 (2007)
Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mouse ProNGF - pro-form of the NGF (nerve growth factor) dimer, 100 kDa Mus musculus protein
|
| Buffer: |
50 mM Na-phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 18
|
Structural and functional properties of mouse proNGF.
Biochem Soc Trans 34(Pt 4):605-6 (2006)
Paoletti F, Konarev PV, Covaceuszach S, Schwarz E, Cattaneo A, Lamba D, Svergun DI
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Hepatocyte growth factor monomer, 79 kDa Mus musculus protein
Hepatocyte growth factor receptor monomer, 100 kDa Mus musculus protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 9
|
Structural basis of hepatocyte growth factor/scatter factor and MET signalling
Proceedings of the National Academy of Sciences 103(11):4046-4051 (2006)
Gherardi E, Sandin S, Petoukhov M, Finch J, Youles M, Ofverstedt L, Miguel R, Blundell T, Vande Woude G, Skoglund U, Svergun D
|
| RgGuinier |
6.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
370 |
nm3 |
|
|