|
|
|
|
|
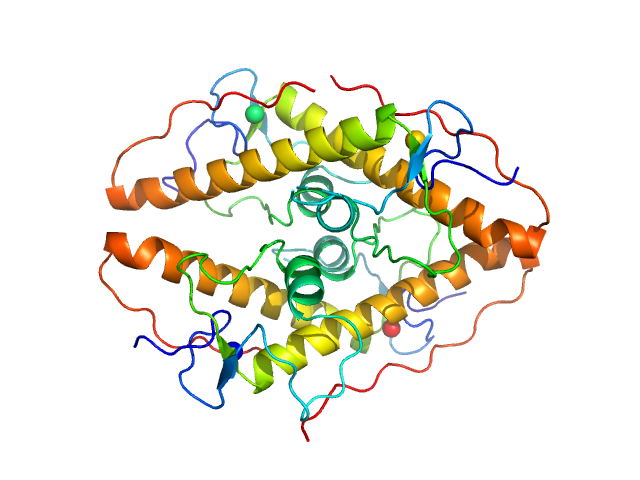
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
| RgGuinier |
5.7 |
nm |
| Dmax |
18.9 |
nm |
|
|
|
|
|
|
|
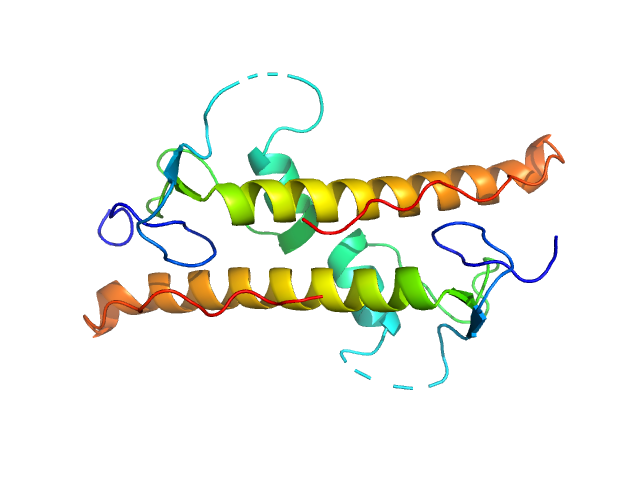
| Sample: |
LD15650p (Pita, isoform A) tetramer, 52 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure (2022)
Bonchuk AN, Boyko KM, Nikolaeva AY, Burtseva AD, Popov VO, Georgiev PG
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
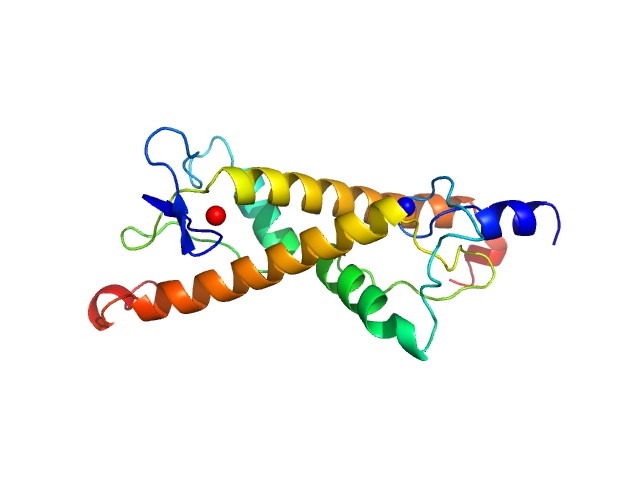
| Sample: |
LD15650p (Pita, isoform A; L45A) dimer, 26 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 17
|
Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure (2022)
Bonchuk AN, Boyko KM, Nikolaeva AY, Burtseva AD, Popov VO, Georgiev PG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LD30467p (Motif 1 binding protein) dimer, 22 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Bis-Tris-Propane, 100 mM NaCl, 5 mM DTT, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 17
|
Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure (2022)
Bonchuk AN, Boyko KM, Nikolaeva AY, Burtseva AD, Popov VO, Georgiev PG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|

|
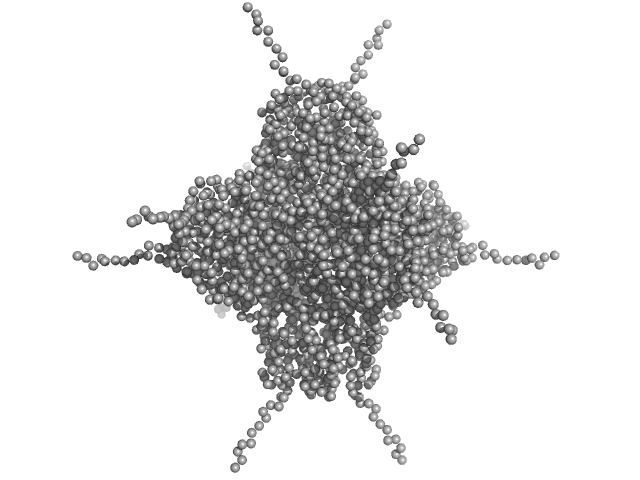
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
409 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 80 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
475 |
nm3 |
|
|
|
|
|
|
|
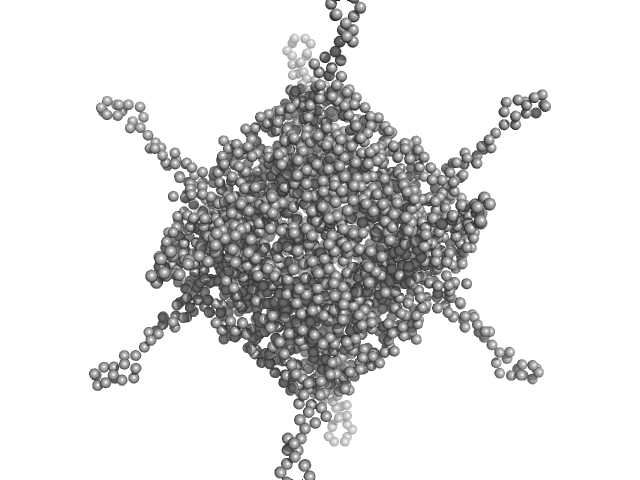
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.5 |
nm |
| Dmax |
20.6 |
nm |
| VolumePorod |
438 |
nm3 |
|
|
|
|
|
|
|
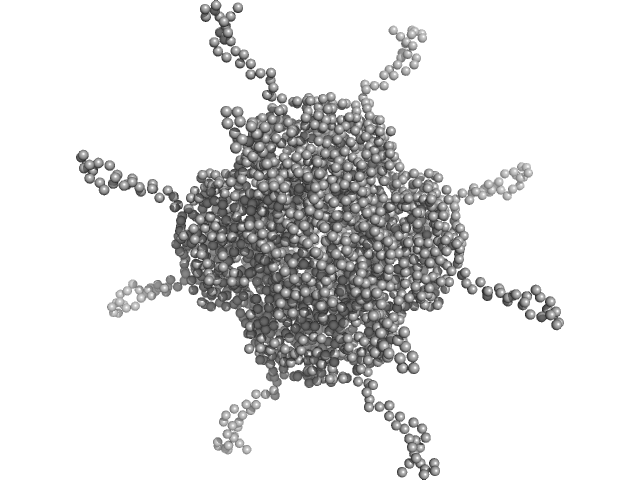
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 480 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
430 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation, DPS-∆N (Ferritin superfamily) dodecamer, 218 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation, DPS-∆N (Ferritin superfamily) dodecamer, 218 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
291 |
nm3 |
|
|