|
|
|
|
|
| Sample: |
Beta-amylase tetramer, 231 kDa Zea mays protein
|
| Buffer: |
50 mM HEPES, 25 mM NaCl, and 0.2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 7
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
615 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Beta-amylase tetramer, 231 kDa Zea mays protein
|
| Buffer: |
50 mM HEPES, 25 mM NaCl, and 0.2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 7
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
5.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
467 |
nm3 |
|
|
|
|
|
|
|
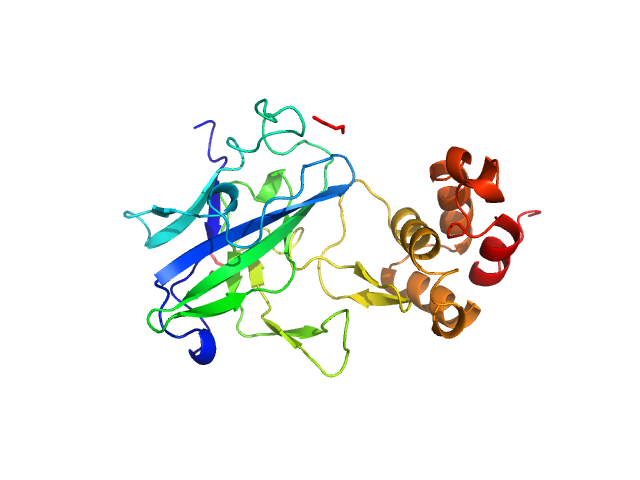
| Sample: |
Beta-amylase tetramer, 224 kDa Ipomoea batatas protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, and 0.2 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 7
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
296 |
nm3 |
|
|
|
|
|
|
|
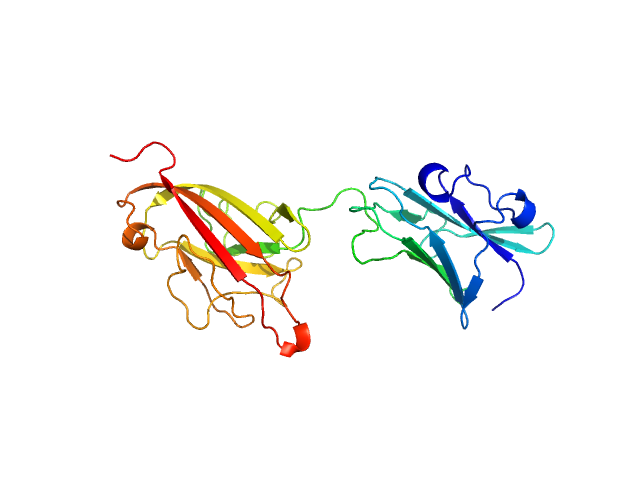
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.1 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
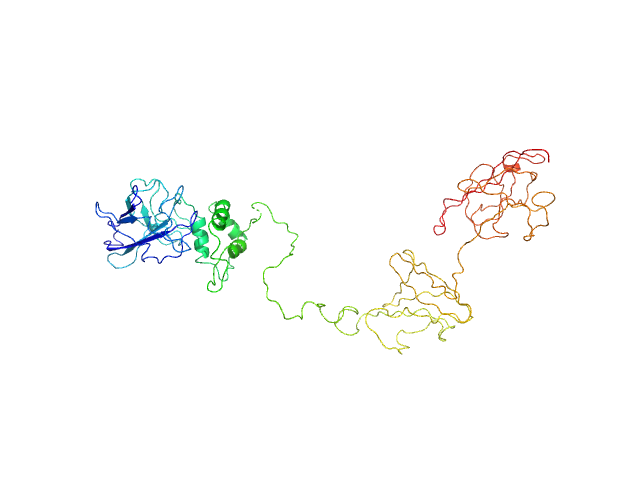
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
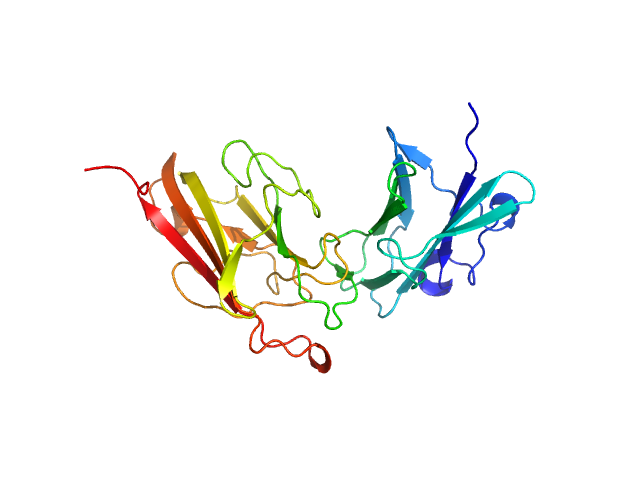
| Sample: |
Dockerin domain-containing protein monomer, 57 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 28
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
5.4 |
nm |
| Dmax |
20.3 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dockerin domain-containing protein monomer, 26 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome
Journal of Biological Chemistry :101896 (2022)
Cerqueira F, Photenhauer A, Doden H, Brown A, Abdel-Hamid A, Moraïs S, Bayer E, Wawrzak Z, Cann I, Ridlon J, Hopkins J, Koropatkin N
|
| RgGuinier |
5.2 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type I restriction-modification system methyltransferase subunit dimer, 144 kDa Vibrio vulnificus (strain … protein
Protein Ocr dimer, 28 kDa Escherichia phage T7 protein
|
| Buffer: |
20 mM Tris-HCl,150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2020 Apr 22
|
Structural features of a minimal intact methyltransferase of a type I restriction-modification system.
Int J Biol Macromol (2022)
Seo PW, Hofmann A, Kim JH, Hwangbo SA, Kim JH, Kim JW, Huynh TYL, Choy HE, Kim SJ, Lee J, Lee JO, Jin KS, Park SY, Kim JS
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
354 |
nm3 |
|
|