|
|
|
|

|
| Sample: |
Circadian clock protein KaiA dodecamer, 393 kDa Synechococcus elongatus (strain … protein
Circadian clock protein KaiB hexamer, 71 kDa Synechococcus elongatus (strain … protein
Circadian clock protein kinase KaiC (S431D mutant) hexamer, 357 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, pH: 7.8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Dec 8
|
Overall structure of fully assembled cyanobacterial KaiABC circadian clock complex by an integrated experimental-computational approach.
Commun Biol 5(1):184 (2022)
Yunoki Y, Matsumoto A, Morishima K, Martel A, Porcar L, Sato N, Yogo R, Tominaga T, Inoue R, Yagi-Utsumi M, Okuda A, Shimizu M, Urade R, Terauchi K, Kono H, Yagi H, Kato K, Sugiyama M
|
| RgGuinier |
7.0 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
1650 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
75% deuterated Circadian clock protein KaiB hexamer, 71 kDa Synechococcus elongatus (strain … protein
Circadian clock protein KaiA dodecamer, 393 kDa Synechococcus elongatus (strain … protein
75% deuterated Circadian clock protein kinase KaiC (S431D mutant) hexamer, 357 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mm NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D2O, pH: 7.8 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Sep 19
|
Overall structure of fully assembled cyanobacterial KaiABC circadian clock complex by an integrated experimental-computational approach.
Commun Biol 5(1):184 (2022)
Yunoki Y, Matsumoto A, Morishima K, Martel A, Porcar L, Sato N, Yogo R, Tominaga T, Inoue R, Yagi-Utsumi M, Okuda A, Shimizu M, Urade R, Terauchi K, Kono H, Yagi H, Kato K, Sugiyama M
|
| RgGuinier |
7.8 |
nm |
| Dmax |
25.6 |
nm |
| VolumePorod |
1620 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Thioredoxin domain-containing protein trimer, 73 kDa Caulobacter vibrioides (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 21
|
The suppressor of copper sensitivity protein C from Caulobacter crescentus
is a trimeric disulfide isomerase that binds copper(I) with subpicomolar affinity
Acta Crystallographica Section D Structural Biology 78(3):337-352 (2022)
Petit G, Hong Y, Djoko K, Whitten A, Furlong E, McCoy A, Gulbis J, Totsika M, Martin J, Halili M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
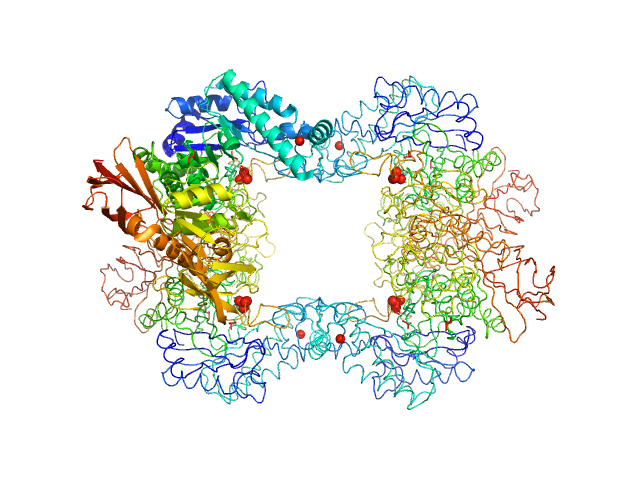
|
| Sample: |
Alpha-L-fucosidase tetramer, 297 kDa Paenibacillus thiaminolyticus (Bacillus … protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2019 Jul 16
|
The first structure–function study of GH151 α‐ l
‐fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
The FEBS Journal (2022)
Koval'ová T, Kovaľ T, Stránský J, Kolenko P, Dušková J, Švecová L, Vodičková P, Spiwok V, Benešová E, Lipovová P, Dohnálek J
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
418 |
nm3 |
|
|
|
|
|
|
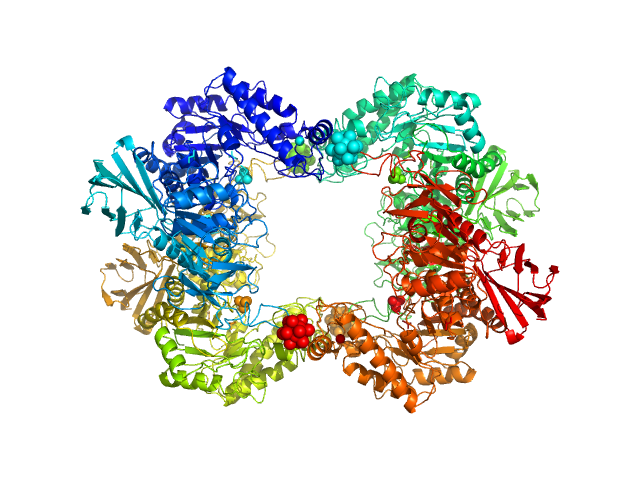
|
| Sample: |
Alpha-L-fucosidase H503A tetramer, 297 kDa Paenibacillus thiaminolyticus (Bacillus … protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7.4 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2019 Jul 16
|
The first structure–function study of GH151 α‐ l
‐fucosidase uncovers new oligomerization pattern, active site complementation, and selective substrate specificity
The FEBS Journal (2022)
Koval'ová T, Kovaľ T, Stránský J, Kolenko P, Dušková J, Švecová L, Vodičková P, Spiwok V, Benešová E, Lipovová P, Dohnálek J
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
433 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
11.2 |
nm |
| Dmax |
37.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
8.6 |
nm |
| Dmax |
34.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
PiPOx239-g-PnPrOx14 monomer, 413 kDa
|
| Buffer: |
ethanol, pH: 7.3 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 12
|
Rigid-to-Flexible Transition in a Molecular Brush in a Good Solvent at a Semidilute Concentration
Langmuir 38(17):5226-5236 (2022)
Kang J, Sachse C, Ko C, Schroer M, Vela S, Molodenskiy D, Kohlbrecher J, Bushuev N, Gumerov R, Potemkin I, Jordan R, Papadakis C
|
| RgGuinier |
7.2 |
nm |
| Dmax |
18.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
200 mM NaCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
RNA triplex structures revealed by WAXS-driven MD simulations
(2022)
Chen Y, He W, Kirmizialtin S, Pollack L
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
|
|
|
|
|
|
|
| Sample: |
RNA Short Tetraloop Hairpin Duplex monomer, 13 kDa RNA
|
| Buffer: |
200 mM KCl, 20 mM Na-MES, 50 μM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2019 Sep 20
|
RNA triplex structures revealed by WAXS-driven MD simulations
(2022)
Chen Y, He W, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.8 |
nm |
|
|