|
|
|
|
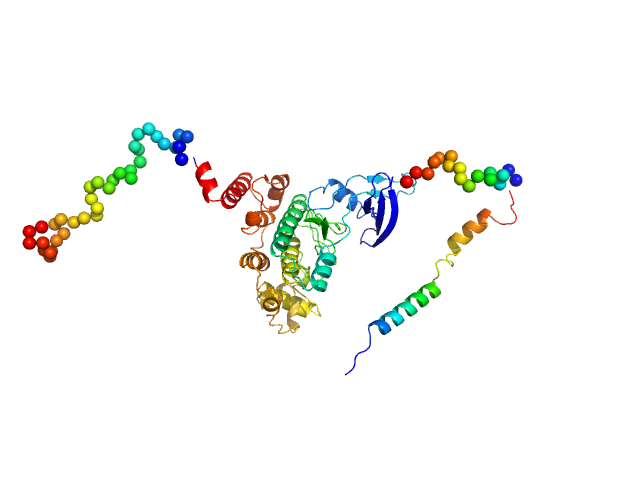
|
| Sample: |
Glycogen synthase kinase 3 monomer, 52 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 19
|
N-terminal phosphorylation regulates the activity of glycogen synthase kinase 3 from Plasmodium falciparum.
Biochem J 479(3):337-356 (2022)
Pazicky S, Alder A, Mertens H, Svergun D, Gilberger T, Löw C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
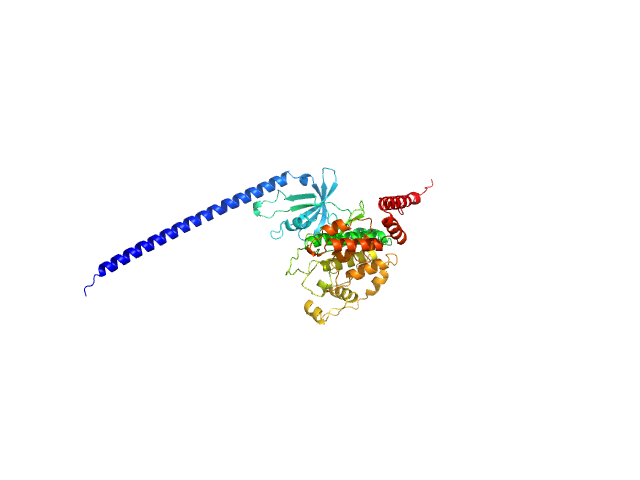
|
| Sample: |
Glycogen synthase kinase 3 monomer, 52 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 19
|
N-terminal phosphorylation regulates the activity of glycogen synthase kinase 3 from Plasmodium falciparum.
Biochem J 479(3):337-356 (2022)
Pazicky S, Alder A, Mertens H, Svergun D, Gilberger T, Löw C
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Outer membrane virulence protein yopE monomer, 15 kDa Vibrio cholerae protein
|
| Buffer: |
10 mM HEPES-HCl, pH 7.4, 150 mM NaCl, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Jun 29
|
Solution structure and dynamics of the mitochondrial‐targeted GTPase
‐activating protein ( GAP
) VopE
by an integrated NMR
/ SAXS
approach
Protein Science (2022)
Smith K, Lee W, Tonelli M, Lee Y, Light S, Cornilescu G, Chakravarthy S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cobalt/magnesium transport protein CorA pentamer, 208 kDa Thermotoga maritima (strain … protein
|
| Buffer: |
20 mM Tris-DCl, 150 mM NaCl, 1 mM EDTA-NaOD, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 23
|
Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation.
Elife 11 (2022)
Johansen NT, Bonaccorsi M, Bengtsen T, Larsen AH, Tidemand FG, Pedersen MC, Huda P, Berndtsson J, Darwish T, Yepuri NR, Martel A, Pomorski TG, Bertarello A, Sansom M, Rapp M, Crehuet R, Schubeis T, Lindorff-Larsen K, Pintacuda G, Arleth L
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
Cobalt/magnesium transport protein CorA pentamer, 208 kDa Thermotoga maritima (strain … protein
|
| Buffer: |
20 mM Tris-DCl, 150 mM NaCl, 40 mM MgCl2, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 23
|
Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation.
Elife 11 (2022)
Johansen NT, Bonaccorsi M, Bengtsen T, Larsen AH, Tidemand FG, Pedersen MC, Huda P, Berndtsson J, Darwish T, Yepuri NR, Martel A, Pomorski TG, Bertarello A, Sansom M, Rapp M, Crehuet R, Schubeis T, Lindorff-Larsen K, Pintacuda G, Arleth L
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.9 |
nm |
|
|
|
|
|
|
|
| Sample: |
Cobalt/magnesium transport protein CorA pentamer, 208 kDa Thermotoga maritima (strain … protein
|
| Buffer: |
20 mM Tris-DCl, 150 mM NaCl, 1 mM EDTA-NaOD, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 23
|
Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation.
Elife 11 (2022)
Johansen NT, Bonaccorsi M, Bengtsen T, Larsen AH, Tidemand FG, Pedersen MC, Huda P, Berndtsson J, Darwish T, Yepuri NR, Martel A, Pomorski TG, Bertarello A, Sansom M, Rapp M, Crehuet R, Schubeis T, Lindorff-Larsen K, Pintacuda G, Arleth L
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Cobalt/magnesium transport protein CorA pentamer, 208 kDa Thermotoga maritima (strain … protein
|
| Buffer: |
20 mM Tris-DCl, 150 mM NaCl, 40 mM MgCl2, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 23
|
Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation.
Elife 11 (2022)
Johansen NT, Bonaccorsi M, Bengtsen T, Larsen AH, Tidemand FG, Pedersen MC, Huda P, Berndtsson J, Darwish T, Yepuri NR, Martel A, Pomorski TG, Bertarello A, Sansom M, Rapp M, Crehuet R, Schubeis T, Lindorff-Larsen K, Pintacuda G, Arleth L
|
| RgGuinier |
4.9 |
nm |
| Dmax |
14.3 |
nm |
|
|
|
|
|
|

|
| Sample: |
5BSL3.2 monomer, 15 kDa Hepatitis C virus … RNA
|
| Buffer: |
10 mM Tris-HCl, 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2019 Jun 2
|
The low-resolution structural models of hepatitis C virus RNA subdomain 5BSL3.2 and its distal complex with domain 3'X point to conserved regulatory mechanisms within the Flaviviridae family.
Nucleic Acids Res (2022)
Castillo-Martínez J, Fan L, Szewczyk MP, Wang YX, Gallego J
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
5BSL3.2 monomer, 15 kDa Hepatitis C virus … RNA
|
| Buffer: |
10 mM Tris-HCl, 6 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2019 Jun 2
|
The low-resolution structural models of hepatitis C virus RNA subdomain 5BSL3.2 and its distal complex with domain 3'X point to conserved regulatory mechanisms within the Flaviviridae family.
Nucleic Acids Res (2022)
Castillo-Martínez J, Fan L, Szewczyk MP, Wang YX, Gallego J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
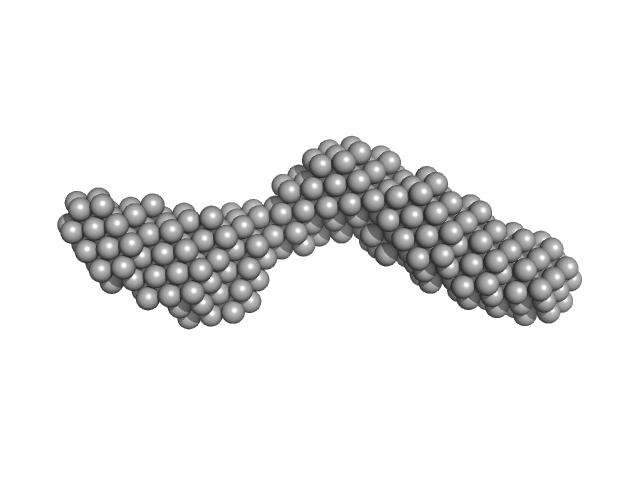
|
| Sample: |
Domain 3'X mutant monomer, 32 kDa Hepatitis C virus … RNA
|
| Buffer: |
10 mM Tris-HCl, 0.1 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, NIDDK, NIH on 2019 Jun 2
|
The low-resolution structural models of hepatitis C virus RNA subdomain 5BSL3.2 and its distal complex with domain 3'X point to conserved regulatory mechanisms within the Flaviviridae family.
Nucleic Acids Res (2022)
Castillo-Martínez J, Fan L, Szewczyk MP, Wang YX, Gallego J
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
70 |
nm3 |
|
|