|
|
|
|
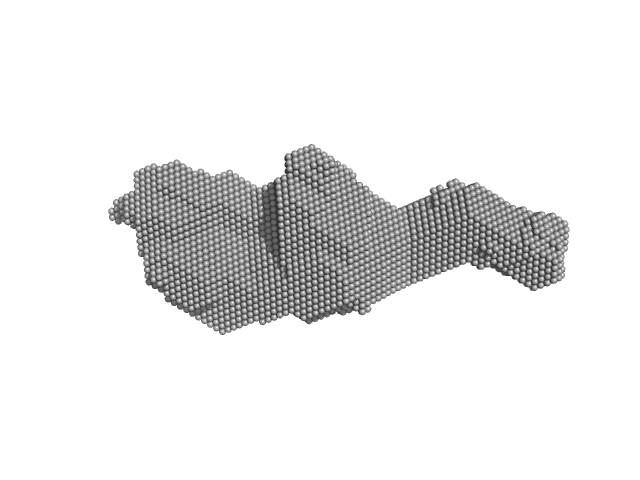
|
| Sample: |
Orange carotenoid-binding protein dimer, 70 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
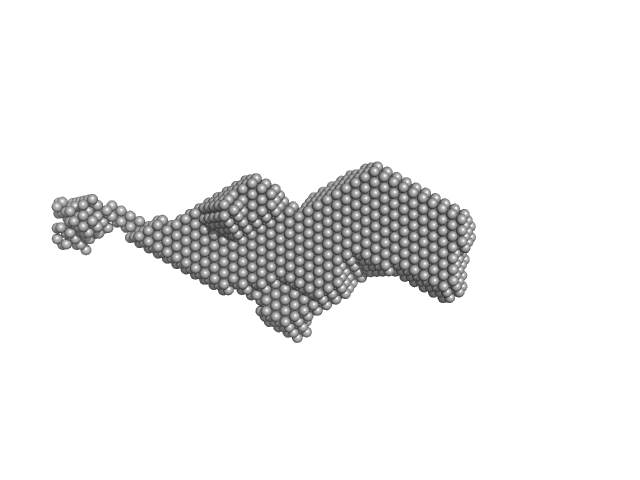
|
| Sample: |
Orange carotenoid-binding protein dimer, 70 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
3.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
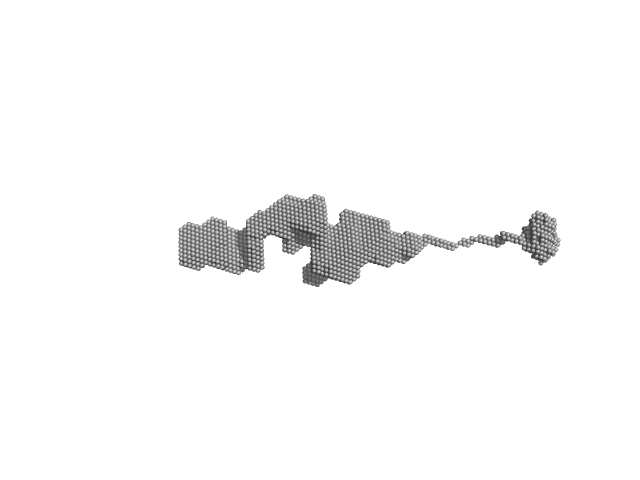
|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
4.4 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
3.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
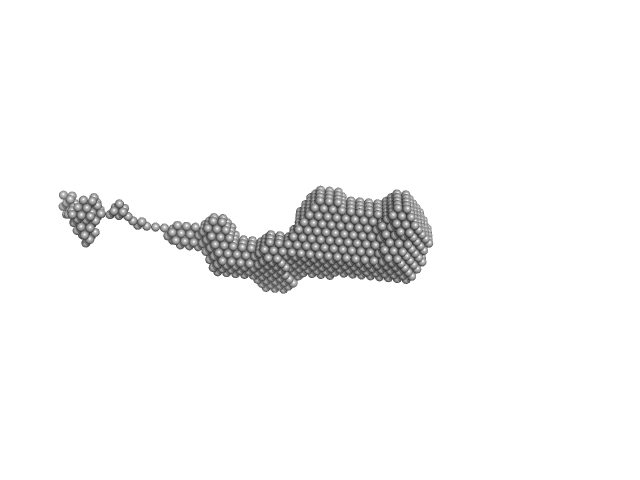
|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
3.4 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
4.8 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Metallothionein-like protein 2 monomer, 8 kDa Cicer arietinum protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 8
|
Structural Characterization of Plant Metallothionein
Dima Molodenskiy
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
5 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Metallothionein-like protein 2 monomer, 8 kDa Cicer arietinum protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl + 1mM TCEP (apoMT), pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 8
|
Structural Characterization of Plant Metallothionein
Dima Molodenskiy
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
The separated apo-linker region peptide from the plant Cicer arietinum monomer, 4 kDa Cicer arietinum protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 8
|
Structural Characterization of Plant Metallothionein
Dima Molodenskiy
|
| RgGuinier |
1.4 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
1 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
A CxxC-linker-CxxC construct binding 1 Cd(II) ion in a CdCys4 site monomer, 5 kDa Cicer arietinum protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 8
|
Structural Characterization of Plant Metallothionein
Dima Molodenskiy
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
|
|