|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
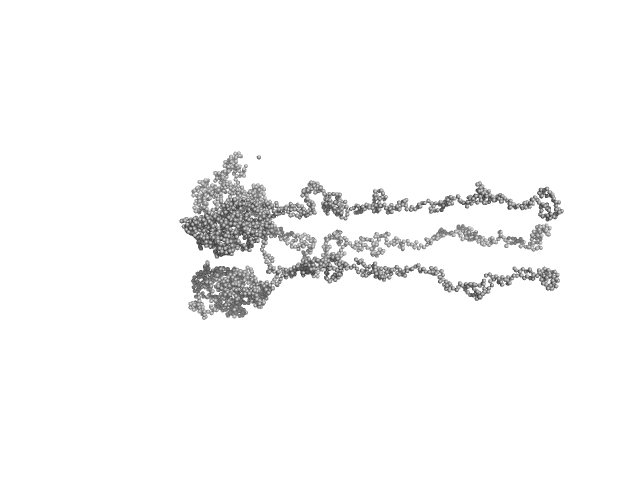
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
8.9 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
1400 mM NaCl, 49.4 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
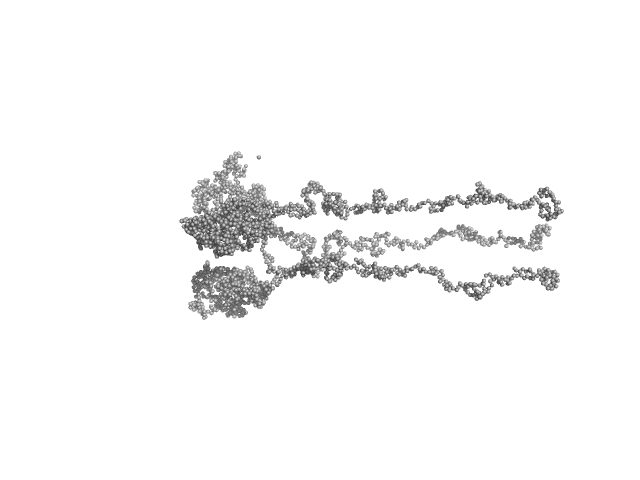
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
8.6 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
2800 mM NaCl, 76.6 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
9.0 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
4000 mM NaCl, 100 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
10.4 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Piwi protein AF_1318 dimer, 98 kDa Archaeoglobus fulgidus protein
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 500 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
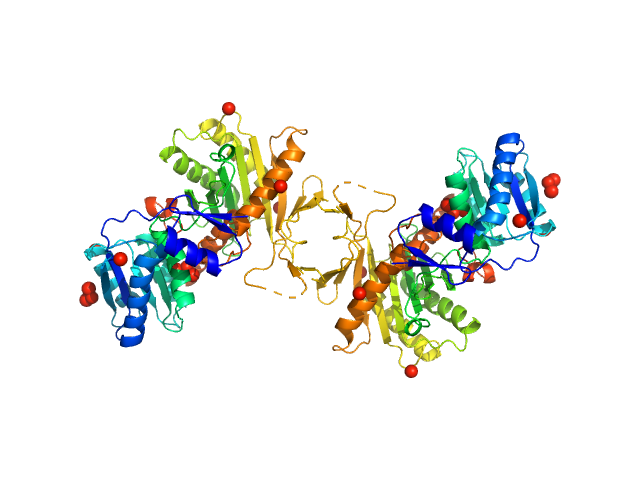
Apo Archaeoglobus fulgidus Argonaute protein dimerises in solution
Elena Manakova
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
174 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMF5_electron_density.png)
|
| Sample: |
Two-component synthetic liposomes composed of 10% DOPS + 90% DOPC lipid mixtures loaded with M1 protein ((lipid:M1 molar ratio 20:1) monomer, 2000 kDa
|
| Buffer: |
100 mM NaCl, 50 mM MES buffer, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Oct 30
|
The Cytoplasmic Tail of Influenza A Virus Hemagglutinin and Membrane Lipid Composition Change the Mode of M1 Protein Association with the Lipid Bilayer
Membranes 11(10):772 (2021)
Kordyukova L, Konarev P, Fedorova N, Shtykova E, Ksenofontov A, Loshkarev N, Dadinova L, Timofeeva T, Abramchuk S, Moiseenko A, Baratova L, Svergun D, Batishchev O
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMG5_electron_density.png)
|
| Sample: |
Two-component synthetic liposomes composed of 30% DOPS + 70% DOPC lipid mixtures loaded with M1 protein (lipid:M1 molar ration 20:1) monomer, 2000 kDa
|
| Buffer: |
100 mM NaCl, 50 mM MES buffer, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Oct 31
|
The Cytoplasmic Tail of Influenza A Virus Hemagglutinin and Membrane Lipid Composition Change the Mode of M1 Protein Association with the Lipid Bilayer
Membranes 11(10):772 (2021)
Kordyukova L, Konarev P, Fedorova N, Shtykova E, Ksenofontov A, Loshkarev N, Dadinova L, Timofeeva T, Abramchuk S, Moiseenko A, Baratova L, Svergun D, Batishchev O
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMH5_electron_density.png)
|
| Sample: |
Four-component synthetic liposomes composed of 30% bPS + 10% POPC + 40% SM + 20% Chol lipid mixtures loaded with M1 protein (lipid:M1 molar ration 20:1) monomer, 2000 kDa
|
| Buffer: |
100 mM NaCl, 50 mM MES buffer, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Oct 31
|
The Cytoplasmic Tail of Influenza A Virus Hemagglutinin and Membrane Lipid Composition Change the Mode of M1 Protein Association with the Lipid Bilayer
Membranes 11(10):772 (2021)
Kordyukova L, Konarev P, Fedorova N, Shtykova E, Ksenofontov A, Loshkarev N, Dadinova L, Timofeeva T, Abramchuk S, Moiseenko A, Baratova L, Svergun D, Batishchev O
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMJ5_electron_density.png)
|
| Sample: |
Four-component synthetic liposomes composed of 20% bPS + 13.3% POPC + 33.3% SM + 33.3% Chol lipid mixtures loaded with M1 protein (lipid:M1 molar ration 20:1) monomer, 2000 kDa
|
| Buffer: |
100 mM NaCl, 50 mM MES buffer, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Oct 31
|
The Cytoplasmic Tail of Influenza A Virus Hemagglutinin and Membrane Lipid Composition Change the Mode of M1 Protein Association with the Lipid Bilayer
Membranes 11(10):772 (2021)
Kordyukova L, Konarev P, Fedorova N, Shtykova E, Ksenofontov A, Loshkarev N, Dadinova L, Timofeeva T, Abramchuk S, Moiseenko A, Baratova L, Svergun D, Batishchev O
|
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDMK5_electron_density.png)
|
| Sample: |
Native liposomes composed of lipids extracted from A/Puerto Rico/8/34 (H1N1) virus envelope loaded with M1 protein. (lipid:M1 molar ration 4:1) monomer, 2000 kDa
|
| Buffer: |
100 mM NaCl, 50 mM MES buffer, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 26
|
The Cytoplasmic Tail of Influenza A Virus Hemagglutinin and Membrane Lipid Composition Change the Mode of M1 Protein Association with the Lipid Bilayer
Membranes 11(10):772 (2021)
Kordyukova L, Konarev P, Fedorova N, Shtykova E, Ksenofontov A, Loshkarev N, Dadinova L, Timofeeva T, Abramchuk S, Moiseenko A, Baratova L, Svergun D, Batishchev O
|
|
|