|
|
|
|

|
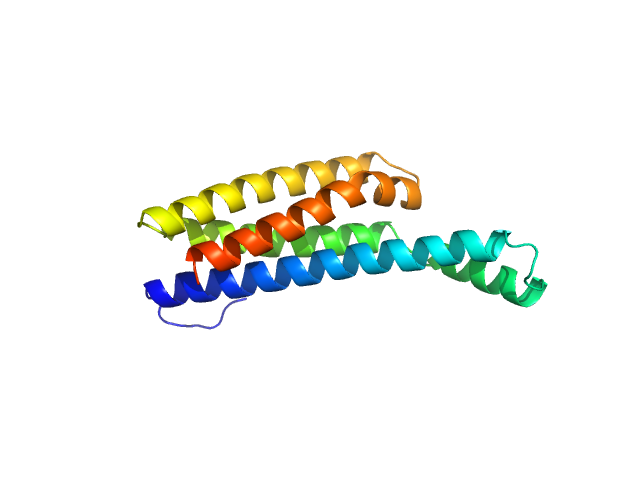
| Sample: |
5-10-5 gapmer phosphorothioate antisense oligonucleotide dimer, 14 kDa
|
| Buffer: |
20 mM Tris-Cl (pH 7.5), 250 mM KCl, 50 mM L-proline, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural basis of dimerization and nucleic acid binding of human DBHS proteins NONO and PSPC1.
Nucleic Acids Res (2021)
Knott GJ, Chong YS, Passon DM, Liang XH, Deplazes E, Conte MR, Marshall AC, Lee M, Fox AH, Bond CS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibronectin-binding protein BBK32 monomer, 17 kDa Borreliella burgdorferi B31 protein
|
| Buffer: |
10 mM HEPES, 140 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 24
|
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
J Immunol 207(11):2856-2867 (2021)
Garrigues RJ, Powell-Pierce AD, Hammel M, Skare JT, Garcia BL
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
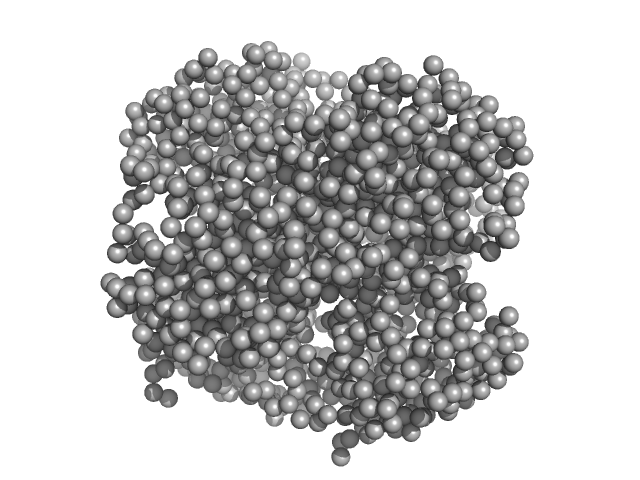
| Sample: |
HpcH/HpaI aldolase hexamer, 165 kDa Rhizorhabdus wittichii RW1 protein
|
| Buffer: |
20 mM HEPES,, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 23
|
EFAMIX
, a tool to decompose inline chromatography SAXS
data from partially overlapping components
Protein Science (2021)
Konarev P, Graewert M, Jeffries C, Fukuda M, Cheremnykh T, Volkov V, Svergun D
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
233 |
nm3 |
|
|
|
|
|
|
|
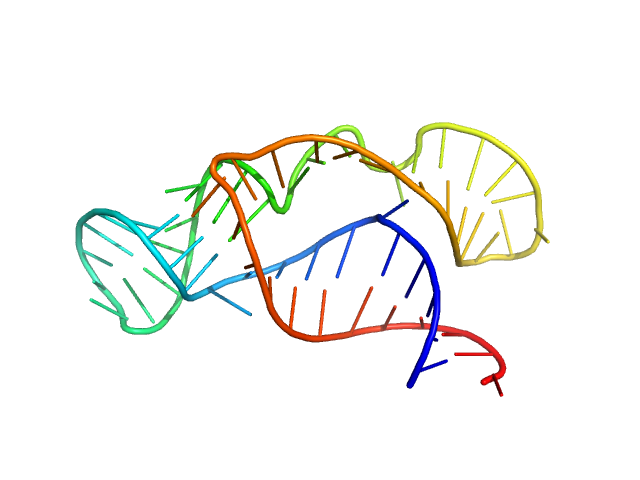
| Sample: |
Neurospora Varkud Satellite ribozyme junction II-III-VI monomer, 20 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 5 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2014 Oct 29
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
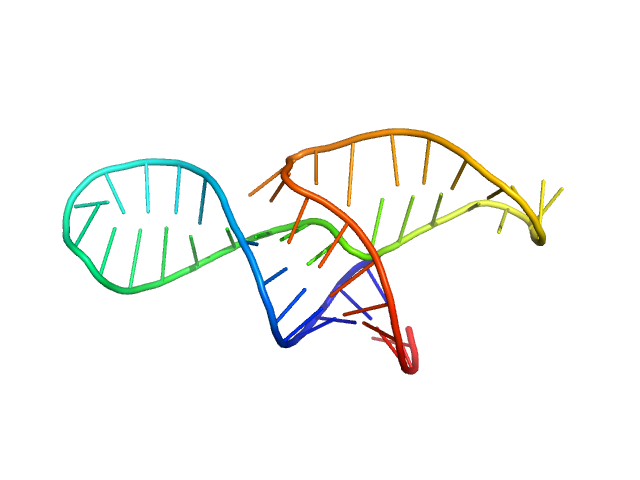
| Sample: |
Neurospora Varkud Satellite ribozyme junction III-IV-V monomer, 15 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 5 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2014 Oct 29
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
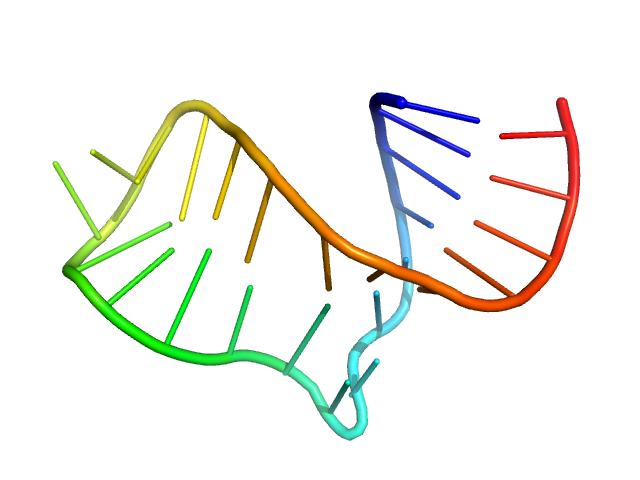
Neurospora Varkud Satellite ribozyme stem-loop VI monomer, 8 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 5 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Jul 7
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
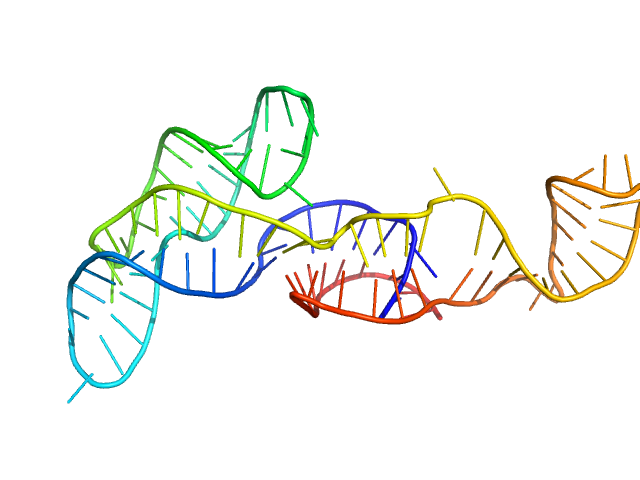
Neurospora Varkud Satellite minimal trans ribozyme monomer, 33 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Sep 19
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurospora Varkud Satellite minimal trans ribozyme monomer, 33 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 5 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Sep 19
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurospora Varkud Satellite minimal trans ribozyme monomer, 33 kDa Neurospora crassa RNA
|
| Buffer: |
50 mM MES, 50 mM KCl, 20 mM MgCl2,, pH: 6.5 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Sep 19
|
An integrative NMR-SAXS approach for structural determination of large RNAs defines the substrate-free state of a trans
-cleaving Neurospora
Varkud Satellite ribozyme
Nucleic Acids Research (2021)
Dagenais P, Desjardins G, Legault P
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
150 mM NaCl, 25 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 8
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
7.6 |
nm |
| Dmax |
41.0 |
nm |
|
|