|
|
|
|
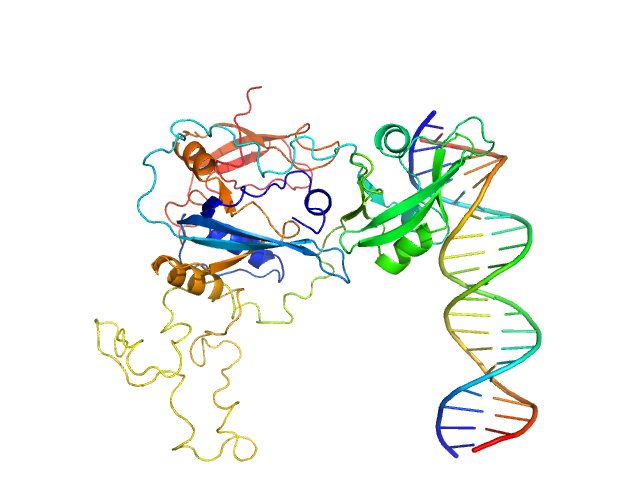
|
| Sample: |
Auxin response factor monomer, 46 kDa Marchantia polymorpha protein
High Affinity ARF binding sequence inverted repeat with 6 nucleotide spacing dimer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2019 Dec 3
|
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Structure (2025)
Crespo I, Malfois M, Rienstra J, Tarrés-Solé A, van den Berg W, Weijers D, Boer DR
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jun 13
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W (artificial PNT3 variant - low-kappa) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jun 13
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.8 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein W (artificial PNT3 variant - high-kappa) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 5
|
Unraveling the molecular grammar and the structural transitions underlying the fibrillation of a viral fibrillogenic domain.
Protein Sci 34(3):e70068 (2025)
Gondelaud F, Leval J, Arora L, Walimbe A, Bignon C, Ptchelkine D, Brocca S, Mukhopadyay S, Longhi S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurotransmitter-gated ion-channel ligand-binding domain-containing protein pentamer, 342 kDa Desulfofustis sp. PB-SRB1 protein
|
| Buffer: |
20 mM citrate, 150 mM NaCl, 10 mM CaCl2, 0.5 mM deuterated n-Dodecyl-B-D-Maltoside, in D2O, pH: 5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2021 Jun 17
|
Structural characterization of pH-modulated closed and open states in a pentameric ligand-gated ion channel
Marie Lycksell
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
581 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurotransmitter-gated ion-channel ligand-binding domain-containing protein pentamer, 342 kDa Desulfofustis sp. PB-SRB1 protein
|
| Buffer: |
20 mM citrate, 150 mM NaCl, 10 mM EDTA, 0.5 mM deuterated n-Dodecyl-β-D-Maltoside, in D2O, pH: 5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2021 Jun 17
|
Structural characterization of pH-modulated closed and open states in a pentameric ligand-gated ion channel
Marie Lycksell
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
558 |
nm3 |
|
|
|
|
|
|
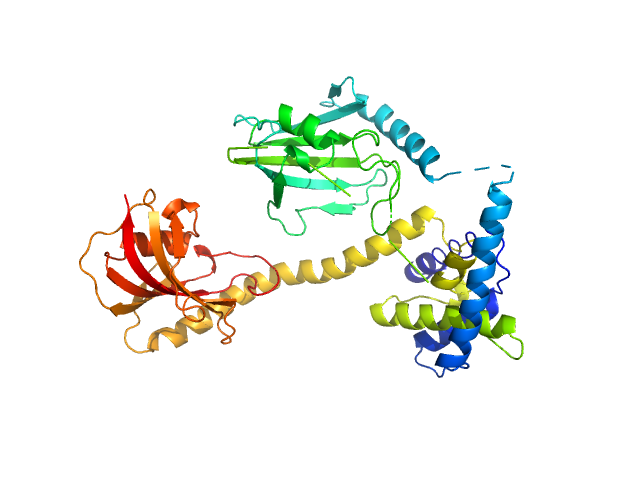
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
(2S)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐4‐methyl‐N‐[(pyridin‐3‐ yl)methyl]pentanamide monomer, 1 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J Med Chem (2025)
Pérez Carrillo VH, Whittaker JJ, Wiedemann C, Harder JM, Lohr T, Jamithireddy AK, Dajka M, Goretzki B, Joseph B, Guskov A, Harmer NJ, Holzgrabe U, Hellmich UA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
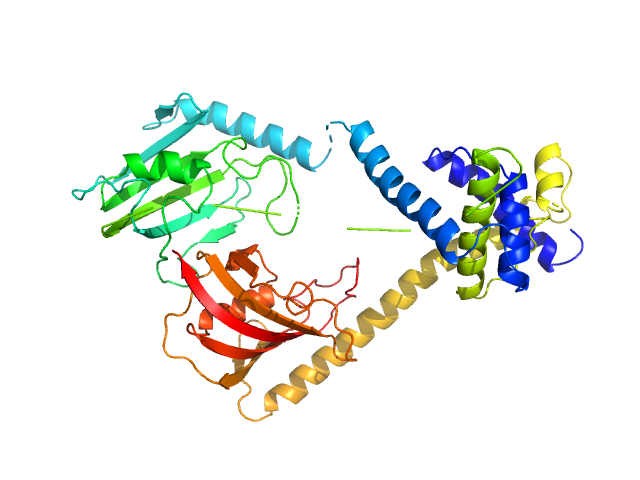
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
(2S)‐3‐(4‐fluorophenyl)‐2‐{[(2S)‐1‐[(4‐ fluorophenyl)methanesulfonyl]piperidin‐2‐ yl]formamido}‐N‐[(pyridin‐3‐yl)methyl]propanamide monomer, 1 kDa
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Structure and Dynamics of Macrophage Infectivity Potentiator Proteins from Pathogenic Bacteria and Protozoans Bound to Fluorinated Pipecolic Acid Inhibitors.
J Med Chem (2025)
Pérez Carrillo VH, Whittaker JJ, Wiedemann C, Harder JM, Lohr T, Jamithireddy AK, Dajka M, Goretzki B, Joseph B, Guskov A, Harmer NJ, Holzgrabe U, Hellmich UA
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|
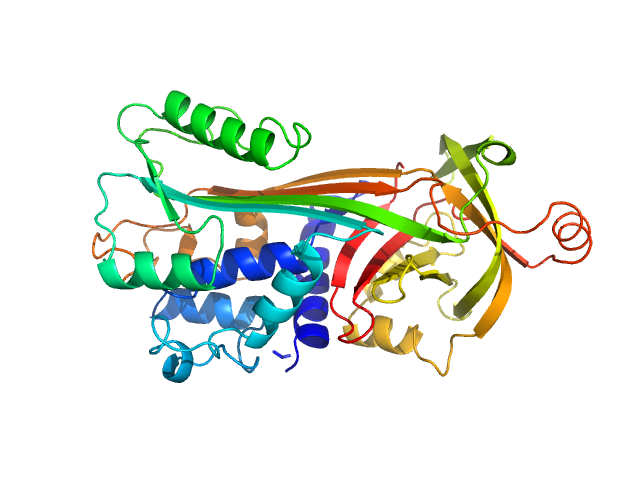
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.5 |
nm |
|
|
|
|
|
|
|
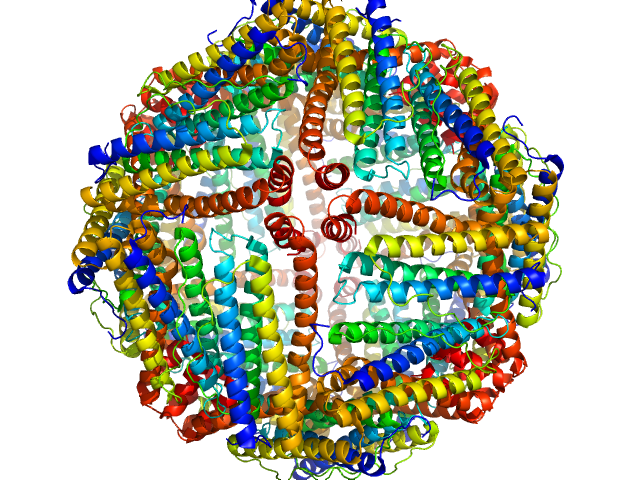
| Sample: |
Ferritin light chain 24-mer, 476 kDa Equus caballus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
23.1 |
nm |
|
|