|
|
|
|
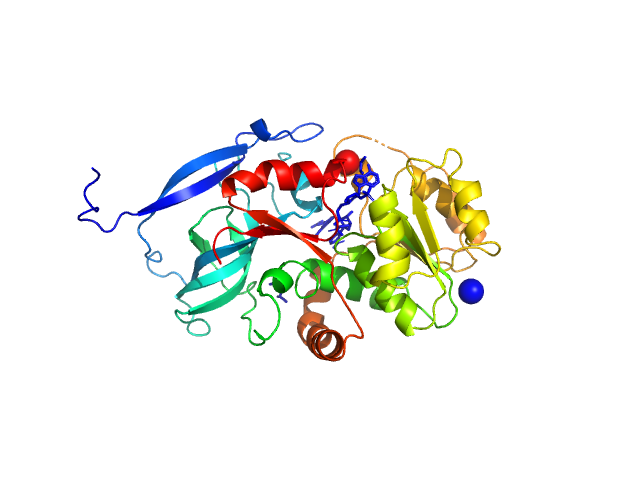
|
| Sample: |
Malus domestica double bond reductase dimer, 77 kDa Malus domestica protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl., pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2020 Oct 23
|
The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates
International Journal of Biological Macromolecules 171:89-99 (2021)
Caliandro R, Polsinelli I, Demitri N, Musiani F, Martens S, Benini S
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
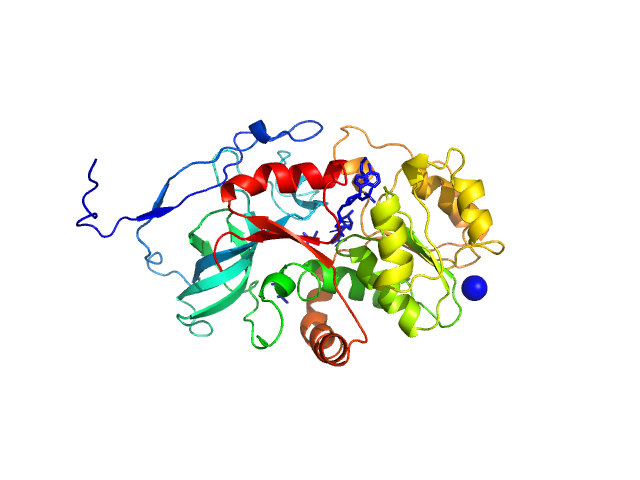
|
| Sample: |
Malus domestica double bond reductase dimer, 77 kDa Malus domestica protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5 mM NADPH, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2020 Oct 23
|
The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates
International Journal of Biological Macromolecules 171:89-99 (2021)
Caliandro R, Polsinelli I, Demitri N, Musiani F, Martens S, Benini S
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Di-domain acyl carrier protein of PigH from prodigiosin biosynthesis monomer, 22 kDa Serratia sp. ATCC … protein
|
| Buffer: |
20 mM Tris supplemented with 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2016 Jan 13
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-hydroxy-2,2'-bipyrrole-5-methanol synthase PigH monomer, 22 kDa Serratia sp. protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
4-hydroxy-2,2'-bipyrrole-5-methanol synthase PigH monomer, 22 kDa Serratia sp. protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 3
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GATA-type iron responsive transcription factor Fep1 reconstituted monomer, 22 kDa Komagataella pastoris protein
24-mer double strand DNA from the GATA promoter dimer, 14 kDa synthetic oligonucleotide DNA
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 5
|
Biophysical characterization of the complex between the iron-responsive transcription factor Fep1 and DNA.
Eur Biophys J (2021)
Miele AE, Cervoni L, Le Roy A, Cutone A, Musci G, Ebel C, Bonaccorsi di Patti MC
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
24-mer double strand DNA from the GATA promoter dimer, 14 kDa synthetic oligonucleotide DNA
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 5
|
Biophysical characterization of the complex between the iron-responsive transcription factor Fep1 and DNA.
Eur Biophys J (2021)
Miele AE, Cervoni L, Le Roy A, Cutone A, Musci G, Ebel C, Bonaccorsi di Patti MC
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GATA-type iron responsive transcription factor Fep1 monomer, 22 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 5
|
Biophysical characterization of the complex between the iron-responsive transcription factor Fep1 and DNA.
Eur Biophys J (2021)
Miele AE, Cervoni L, Le Roy A, Cutone A, Musci G, Ebel C, Bonaccorsi di Patti MC
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GATA-type iron responsive transcription factor Fep1 monomer, 22 kDa Komagataella pastoris protein
24-mer double strand DNA from the GATA promoter dimer, 14 kDa synthetic oligonucleotide DNA
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 5
|
Biophysical characterization of the complex between the iron-responsive transcription factor Fep1 and DNA.
Eur Biophys J (2021)
Miele AE, Cervoni L, Le Roy A, Cutone A, Musci G, Ebel C, Bonaccorsi di Patti MC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
GATA-type iron responsive transcription factor Fep1 reconstituted monomer, 22 kDa Komagataella pastoris protein
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Dec 5
|
Biophysical characterization of the complex between the iron-responsive transcription factor Fep1 and DNA.
Eur Biophys J (2021)
Miele AE, Cervoni L, Le Roy A, Cutone A, Musci G, Ebel C, Bonaccorsi di Patti MC
|
| RgGuinier |
3.8 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|