|
|
|
|
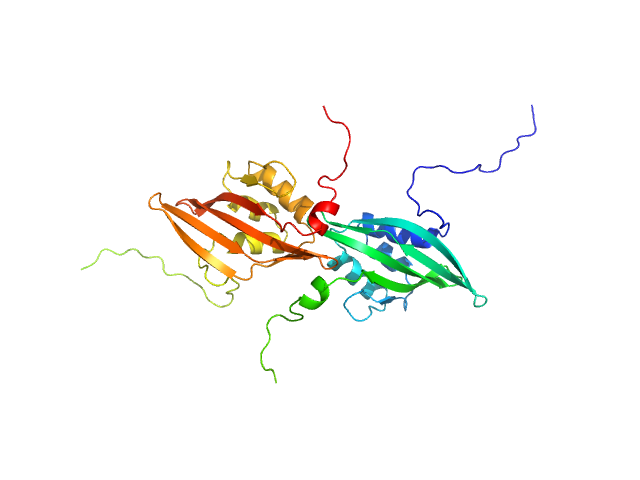
|
| Sample: |
C-terminal domain-like carotenoid protein dimer, 33 kDa Tolypothrix sp. PCC … protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 15
|
Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP
Scientific Reports 10(1) (2020)
Dominguez-Martin M, Hammel M, Gupta S, Lechno-Yossef S, Sutter M, Rosenberg D, Chen Y, Petzold C, Ralston C, Polívka T, Kerfeld C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
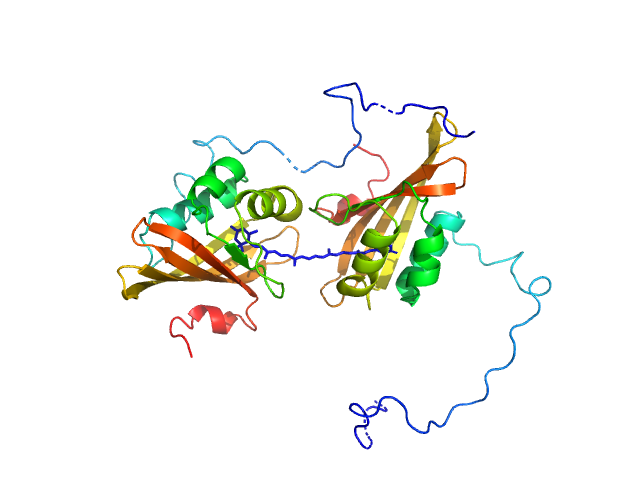
|
| Sample: |
C-terminal domain-like carotenoid protein dimer, 36 kDa Tolypothrix sp. PCC … protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 15
|
Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP
Scientific Reports 10(1) (2020)
Dominguez-Martin M, Hammel M, Gupta S, Lechno-Yossef S, Sutter M, Rosenberg D, Chen Y, Petzold C, Ralston C, Polívka T, Kerfeld C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
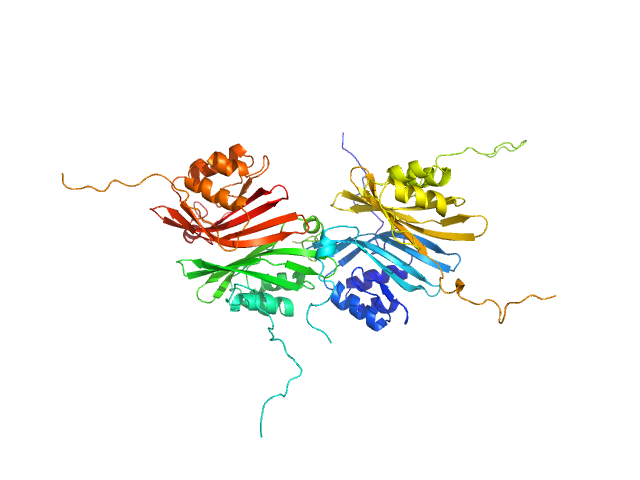
|
| Sample: |
C-terminal domain-like carotenoid protein tetramer, 66 kDa Tolypothrix sp. PCC … protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 15
|
Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP
Scientific Reports 10(1) (2020)
Dominguez-Martin M, Hammel M, Gupta S, Lechno-Yossef S, Sutter M, Rosenberg D, Chen Y, Petzold C, Ralston C, Polívka T, Kerfeld C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
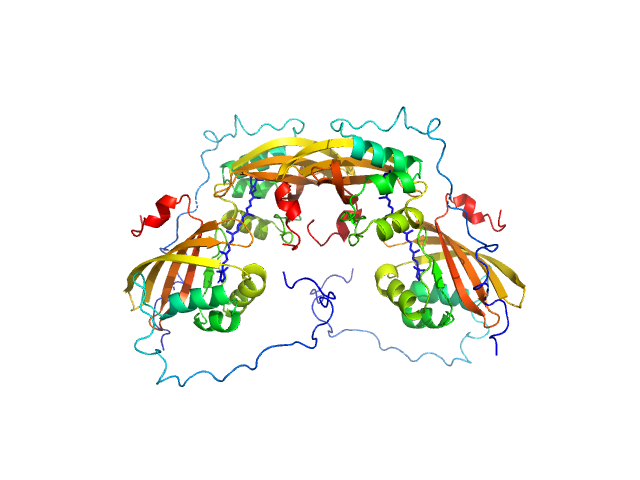
|
| Sample: |
C-terminal domain-like carotenoid protein tetramer, 73 kDa Tolypothrix sp. PCC … protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 15
|
Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP
Scientific Reports 10(1) (2020)
Dominguez-Martin M, Hammel M, Gupta S, Lechno-Yossef S, Sutter M, Rosenberg D, Chen Y, Petzold C, Ralston C, Polívka T, Kerfeld C
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
|
|
|
|
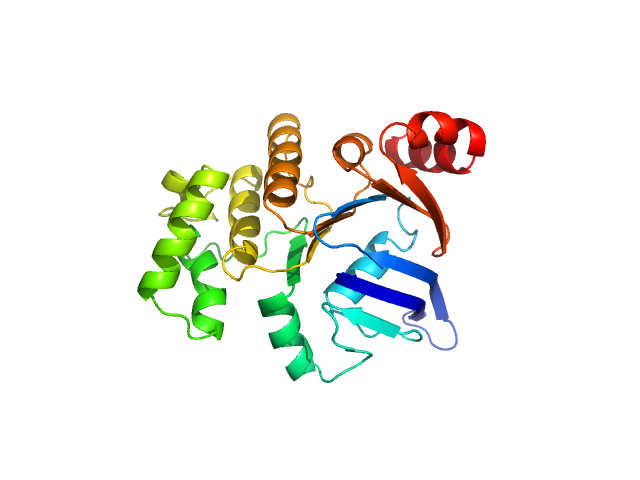
|
| Sample: |
ABC transporter, ATP-binding protein (Nucleotide-Binding Domain SaNsrF) monomer, 31 kDa Streptococcus agalactiae protein
|
| Buffer: |
100 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Dec 11
|
Characterization of the nucleotide-binding domain NsrF from the BceAB-type ABC-transporter NsrFP from the human pathogen Streptococcus agalactiae
Scientific Reports 10(1) (2020)
Furtmann F, Porta N, Hoang D, Reiners J, Schumacher J, Gottstein J, Gohlke H, Smits S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleotide pyrophosphatase/phosphodiesterase from E. characias latex partially sequenced dimer, 28 kDa Euphorbia characias protein
|
| Buffer: |
50 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Feb 20
|
Structure of a nucleotide pyrophosphatase/phosphodiesterase (NPP) from Euphorbia characias latex characterized by small-angle X-ray scattering: clues for the general organization of plant NPPs.
Acta Crystallogr D Struct Biol 76(Pt 9):857-867 (2020)
Sabatucci A, Pintus F, Cabras T, Vincenzoni F, Maccarrone M, Medda R, Dainese E
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
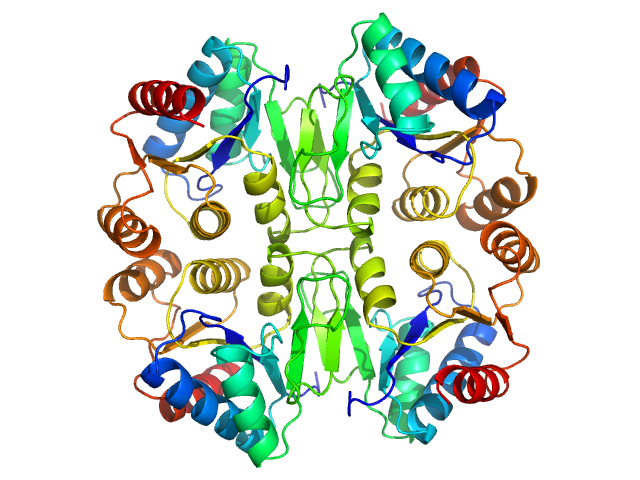
|
| Sample: |
P450 cytochrome, putative (Moco carrier protein) tetramer, 75 kDa Rippkaea orientalis (strain … protein
|
| Buffer: |
100 mM Tris-HCl, 300 mM NaCl, 2 %(v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Feb 5
|
The structure of the Moco carrier protein from Rippkaea orientalis.
Acta Crystallogr F Struct Biol Commun 76(Pt 9):453-463 (2020)
Krausze J, Hercher TW, Archna A, Kruse T
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type 3 secretion system pilotin dimer, 28 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Mar 23
|
Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure (2020)
Majewski DD, Okon M, Heinkel F, Robb CS, Vuckovic M, McIntosh LP, Strynadka NCJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type 3 secretion system pilotin dimer, 19 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Mar 23
|
Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure (2020)
Majewski DD, Okon M, Heinkel F, Robb CS, Vuckovic M, McIntosh LP, Strynadka NCJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 57 kDa Bordetella pertussis protein
|
| Buffer: |
10 mM Tris HCl, 150 mM NaCl, 10 mM CaCl₂, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
Continuous Assembly of β-Roll Structures Is Implicated in the Type I-Dependent Secretion of Large Repeat-in-Toxins (RTX) Proteins.
J Mol Biol (2020)
Motlova L, Klimova N, Fiser R, Sebo P, Bumba L
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|