|
|
|
|
|
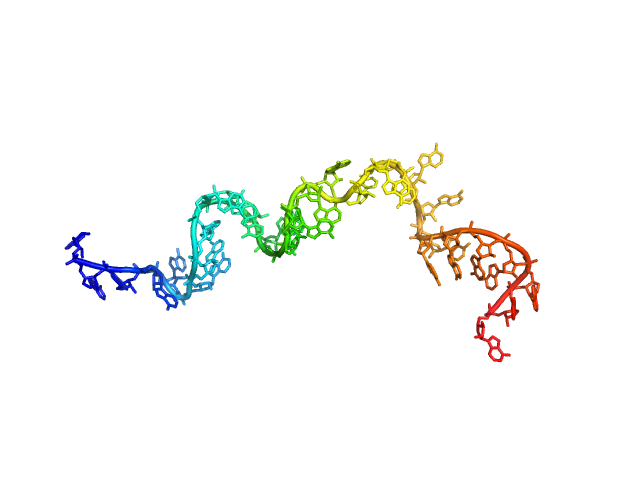
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 600 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 1 mM MgCl2, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2019 Jul 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 2 mM MgCl2, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 5 mM MgCl2, 20µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|

|
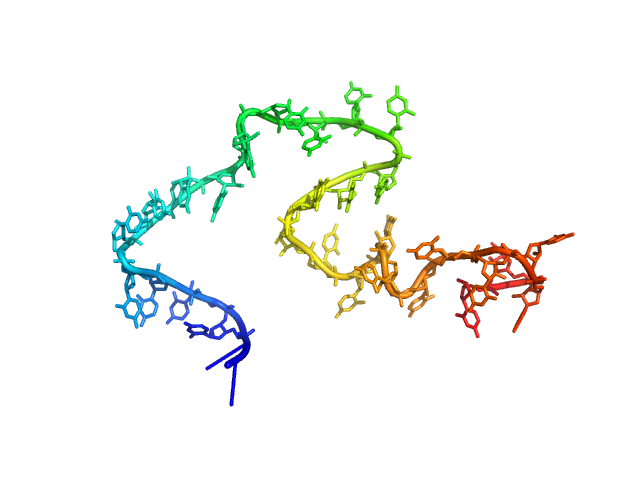
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 100 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 200 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 400 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
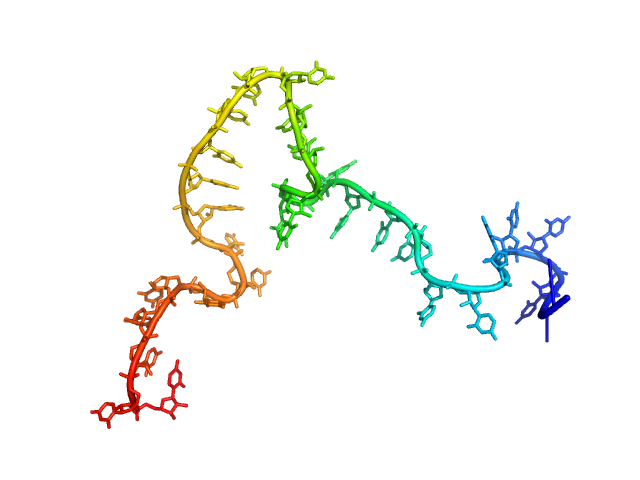
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 600 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 1 mM MgCl2, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|