|
|
|
|
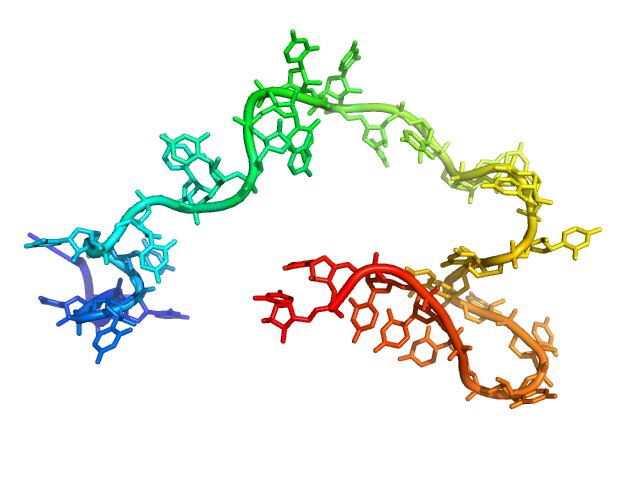
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 2 mM MgCl2, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 5 mM MgCl2, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 10 mM MgCl2, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
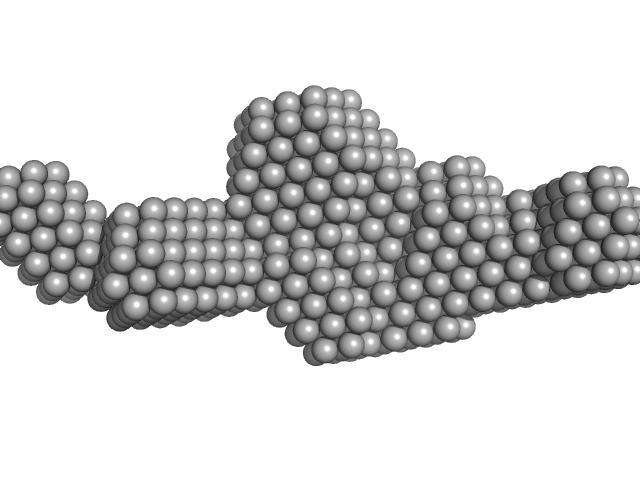
|
| Sample: |
Sa0446 binding sequence 40bp monomer, 25 kDa DNA
Transcriptional regulator Lrs14-like protein dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
300 mM NaCl, 20 mM HEPES, pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 5
|
Solution Structure of Archaeal Biofilm Regulator 2 (AbfR2) in Complex with 40 bp DNA
Marian Vogt
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 248 kDa Yersinia pestis protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
470 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 256 kDa Francisella tularensis protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
491 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 250 kDa Burkholderia pseudomallei protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
437 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calredoxin, Redox protein from Chlamydomonas reinhardtii monomer, 40 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, 5 mM EGTA, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Structural Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University on 2015 Nov 17
|
Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green alga Chlamydomonas reinhardtii.
J Biol Chem (2019)
Charoenwattanasatien R, Zinzius K, Scholz M, Wicke S, Tanaka H, Brandenburg JS, Marchetti GM, Ikegami T, Matsumoto T, Oda T, Sato M, Hippler M, Kurisu G
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Calredoxin, Redox protein from Chlamydomonas reinhardtii monomer, 40 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, 5 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Structural Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University on 2015 Nov 17
|
Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green alga Chlamydomonas reinhardtii.
J Biol Chem (2019)
Charoenwattanasatien R, Zinzius K, Scholz M, Wicke S, Tanaka H, Brandenburg JS, Marchetti GM, Ikegami T, Matsumoto T, Oda T, Sato M, Hippler M, Kurisu G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human dystrophin central domain R8-15 fragment monomer, 100 kDa protein
|
| Buffer: |
NaP 10 mM, NaCl 500 mM, EDTA 1 mM, Glycerol 2%, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Sep 23
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
10.1 |
nm |
| Dmax |
36.0 |
nm |
|
|