|
|
|
|
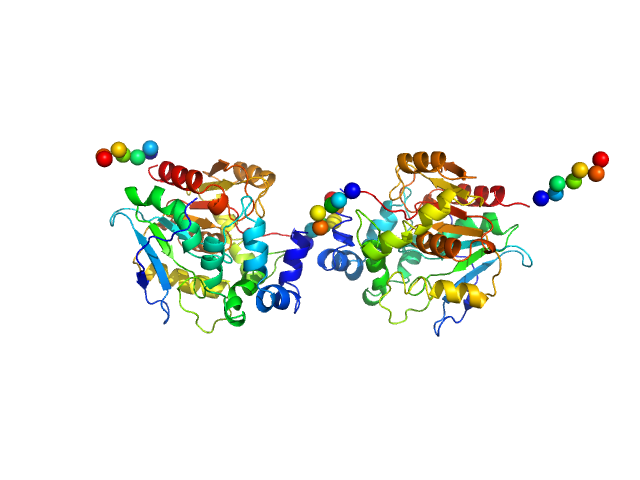
|
| Sample: |
Haloalkane dehalogenase variant DhaA115 - dimeric fraction dimer, 69 kDa Rhodococcus rhodochrous protein
|
| Buffer: |
50 mM potassium phosphate buffer (41 mM K₂HPO₄, 9mM KH₂PO₄), pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2019 Aug 22
|
Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst
Chemical Science 11(41):11162-11178 (2020)
Markova K, Chmelova K, Marques S, Carpentier P, Bednar D, Damborsky J, Marek M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
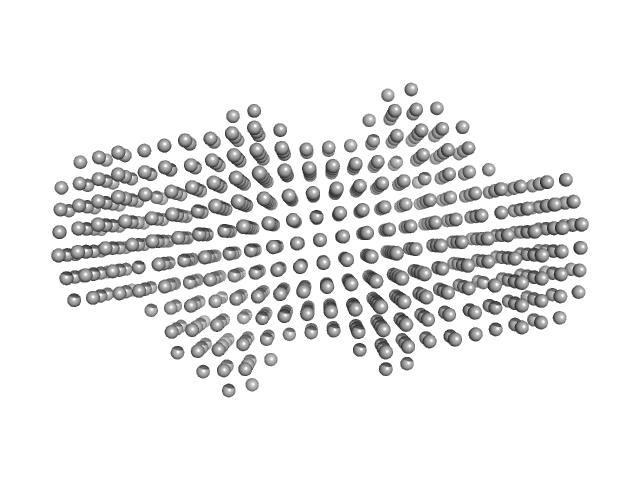
|
| Sample: |
P. maculata perivitellin 2 dimer, 188 kDa Pomacea maculata protein
|
| Buffer: |
20 mM Tris, pH: 7 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 Mar 26
|
Exaptation of two ancient immune proteins into a new dimeric pore-forming toxin in snails.
J Struct Biol 211(2):107531 (2020)
Giglio ML, Ituarte S, Milesi V, Dreon MS, Brola TR, Caramelo J, Ip JCH, Maté S, Qiu JW, Otero LH, Heras H
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
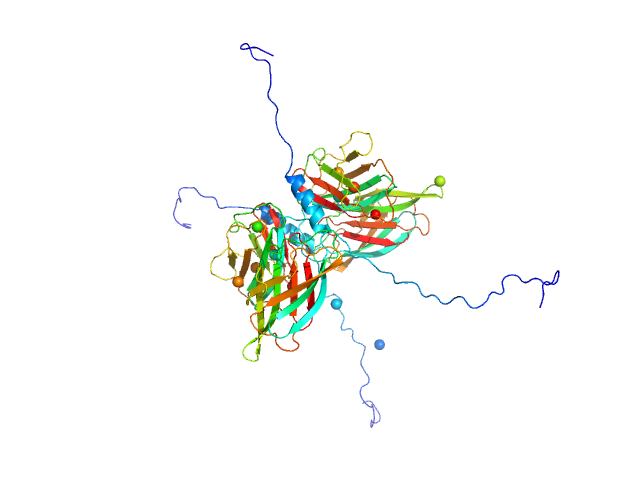
|
| Sample: |
Conserved flagellar protein F dimer, 32 kDa Sulfolobus acidocaldarius protein
Stator protein FlaG soluble domain dimer, 30 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
25 mM citric acid/sodium citrate, 150mM NaCl, 3% Glycerol, pH: 3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 10
|
The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility.
Nat Microbiol (2019)
Tsai CL, Tripp P, Sivabalasarma S, Zhang C, Rodriguez-Franco M, Wipfler RL, Chaudhury P, Banerjee A, Beeby M, Whitaker RJ, Tainer JA, Albers SV
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
109 |
nm3 |
|
|
|
|
|
|
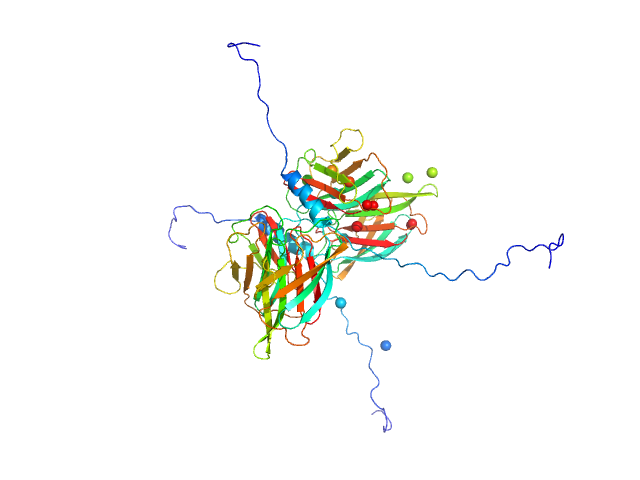
|
| Sample: |
Conserved flagellar protein F dimer, 32 kDa Sulfolobus acidocaldarius protein
Stator protein FlaG-V118K soluble domain dimer, 30 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
25 mM citric acid/sodium citrate, 150mM NaCl, 3% Glycerol, pH: 3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 10
|
The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility.
Nat Microbiol (2019)
Tsai CL, Tripp P, Sivabalasarma S, Zhang C, Rodriguez-Franco M, Wipfler RL, Chaudhury P, Banerjee A, Beeby M, Whitaker RJ, Tainer JA, Albers SV
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Conserved flagellar protein FlaG soluble domain monomer, 15 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
25 mM citric acid/sodium citrate, 150mM NaCl, 3% Glycerol, pH: 3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 10
|
The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility.
Nat Microbiol (2019)
Tsai CL, Tripp P, Sivabalasarma S, Zhang C, Rodriguez-Franco M, Wipfler RL, Chaudhury P, Banerjee A, Beeby M, Whitaker RJ, Tainer JA, Albers SV
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
133 |
nm3 |
|
|
|
|
|
|
|
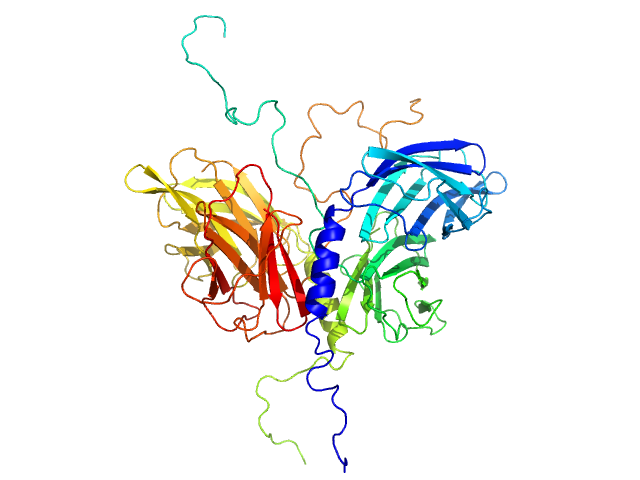
| Sample: |
Stator protein FlaG soluble domain dimer, 30 kDa Sulfolobus acidocaldarius protein
Conserved flagellar protein FlaF-I96Y soluble domain dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
25 mM citric acid/sodium citrate, 150mM NaCl, 3% Glycerol, pH: 3 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 10
|
The structure of the periplasmic FlaG-FlaF complex and its essential role for archaellar swimming motility.
Nat Microbiol (2019)
Tsai CL, Tripp P, Sivabalasarma S, Zhang C, Rodriguez-Franco M, Wipfler RL, Chaudhury P, Banerjee A, Beeby M, Whitaker RJ, Tainer JA, Albers SV
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
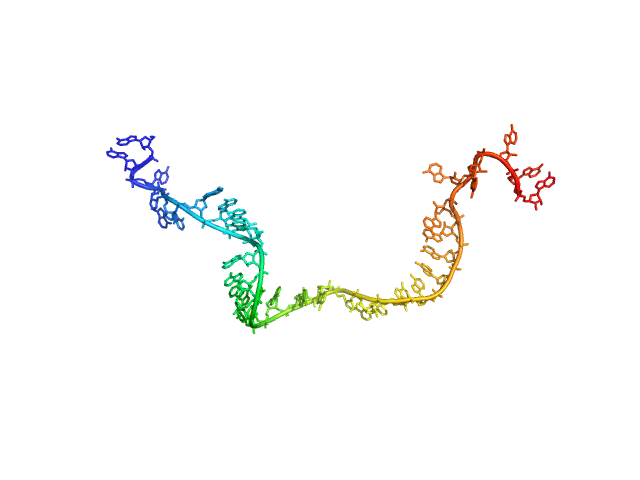
|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
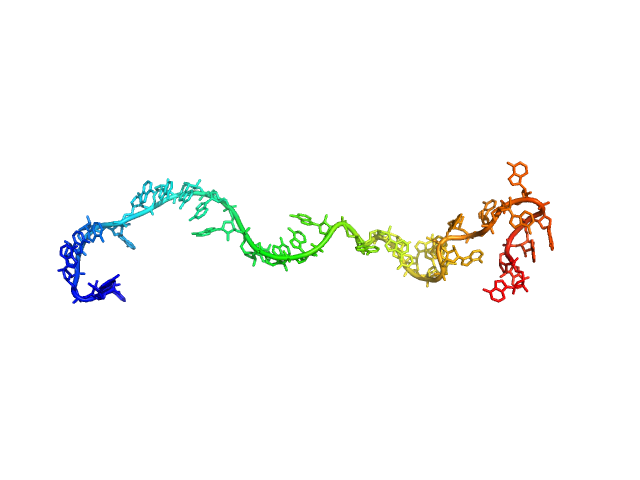
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 100 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
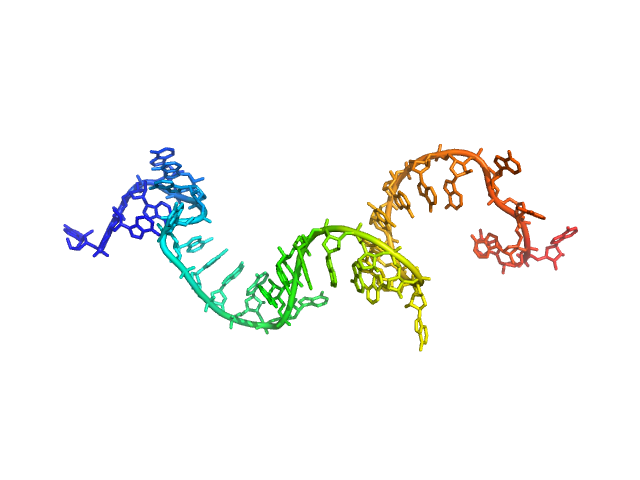
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 200 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-adenosine monomer, 10 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 400 mM NaCl, 20 µM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 25
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|