|
|
|
|
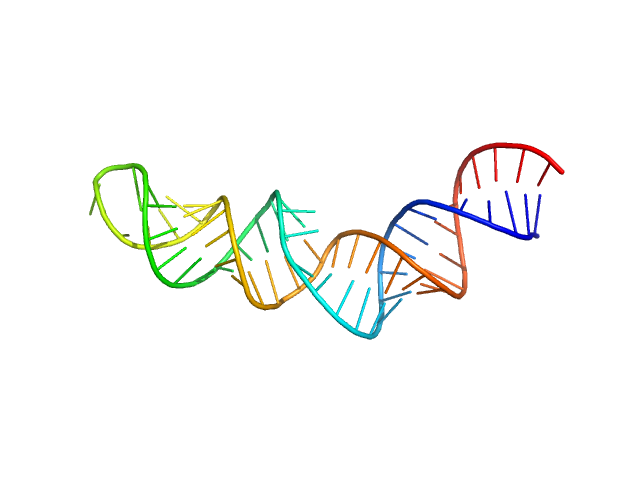
|
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
HIV-1 DIS with a GGGGGG apical loop, UCU bulge, and 16 bp helical extension monomer, 21 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at CG-3, High Flux Isotope Reactor on 2023 Sep 15
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Optimization of Structure‐Guided Development of Chemical Probes for the Pseudoknot RNA of the Frameshift Element in SARS‐CoV‐2
Angewandte Chemie International Edition (2025)
Ceylan B, Adam J, Toews S, Kaiser F, Dörr J, Scheppa D, Tants J, Smart A, Schoth J, Philipp S, Stirnal E, Ferner J, Richter C, Sreeramulu S, Caliskan N, Schlundt A, Weigand J, Göbel M, Wacker A, Schwalbe H
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Optimization of Structure‐Guided Development of Chemical Probes for the Pseudoknot RNA of the Frameshift Element in SARS‐CoV‐2
Angewandte Chemie International Edition (2025)
Ceylan B, Adam J, Toews S, Kaiser F, Dörr J, Scheppa D, Tants J, Smart A, Schoth J, Philipp S, Stirnal E, Ferner J, Richter C, Sreeramulu S, Caliskan N, Schlundt A, Weigand J, Göbel M, Wacker A, Schwalbe H
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SARS-CoV2 RNA pseudoknot monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 50 mM KCl, pH: 6.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
Optimization of Structure‐Guided Development of Chemical Probes for the Pseudoknot RNA of the Frameshift Element in SARS‐CoV‐2
Angewandte Chemie International Edition (2025)
Ceylan B, Adam J, Toews S, Kaiser F, Dörr J, Scheppa D, Tants J, Smart A, Schoth J, Philipp S, Stirnal E, Ferner J, Richter C, Sreeramulu S, Caliskan N, Schlundt A, Weigand J, Göbel M, Wacker A, Schwalbe H
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
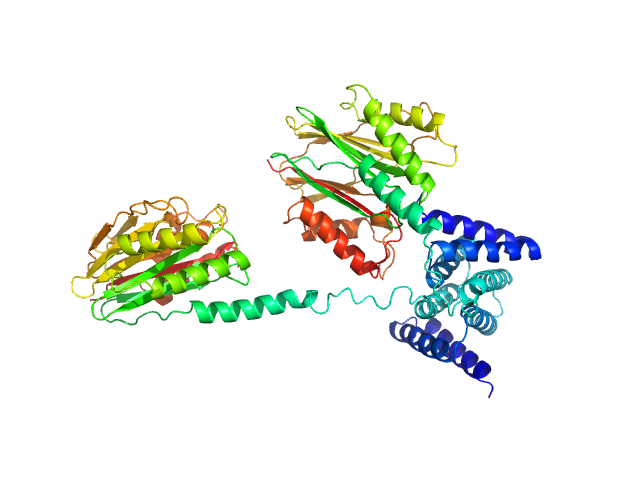
|
| Sample: |
Phosphoserine phosphatase RsbU dimer, 77 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 24
|
A general mechanism for initiating the bacterial general stress response.
Elife 13 (2025)
Baral R, Ho K, Kumar RP, Hopkins JB, Watkins MB, LaRussa S, Caban-Penix S, Calderone LA, Bradshaw N
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Serine/threonine-protein kinase RsbT dimer, 29 kDa Bacillus subtilis (strain … protein
Phosphoserine phosphatase RsbU (Q94L) dimer, 77 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 24
|
A general mechanism for initiating the bacterial general stress response.
Elife 13 (2025)
Baral R, Ho K, Kumar RP, Hopkins JB, Watkins MB, LaRussa S, Caban-Penix S, Calderone LA, Bradshaw N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
119 |
nm3 |
|
|
|
|
|
|
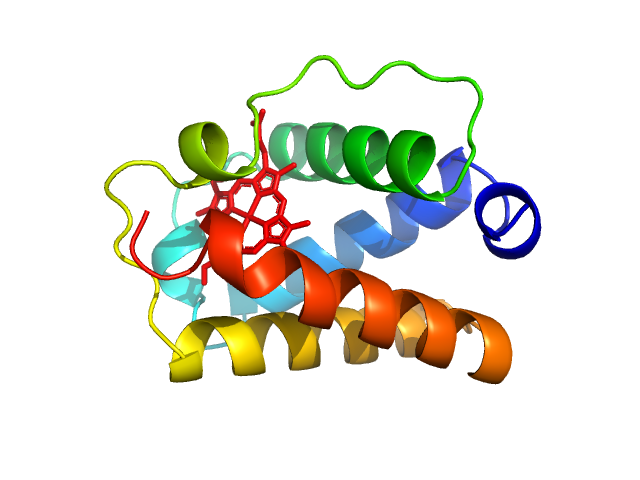
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 May 1
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|