|
|
|
|
|
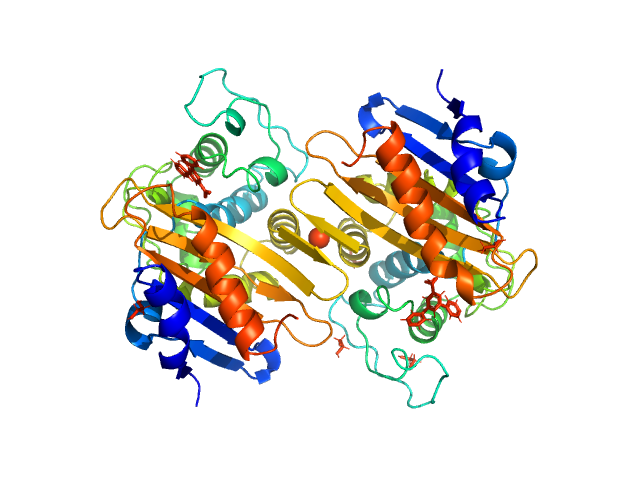
| Sample: |
Beta-lactamase dimer, 56 kDa Klebsiella pneumoniae protein
|
| Buffer: |
50 mM HEPES 50 mM K2SO4, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 23
|
The biological assembly of OXA-48 reveals a dimer interface with high charge complementarity and very high affinity.
FEBS J (2018)
Lund BA, Thomassen AM, Nesheim BHB, Carlsen TJO, Isaksson J, Christopeit T, Leiros HS
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
MOPS map2c buffer, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
|
|
|
|
|
| Sample: |
Truncated P5abc subdomain from tetrahymena ribozyme monomer, 18 kDa RNA
|
| Buffer: |
10mM KMOPS 20mM KCl 1mM MgCl2 20uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 3
|
Revealing the distinct folding phases of an RNA three-helix junction.
Nucleic Acids Res 46(14):7354-7365 (2018)
Plumridge A, Katz AM, Calvey GD, Elber R, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Truncated P5abc subdomain from tetrahymena ribozyme monomer, 18 kDa RNA
|
| Buffer: |
10mM KMOPS 20mM KCl 1mM MgCl2 20uM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Dec 3
|
Revealing the distinct folding phases of an RNA three-helix junction.
Nucleic Acids Res 46(14):7354-7365 (2018)
Plumridge A, Katz AM, Calvey GD, Elber R, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
|
|