|
|
|
|
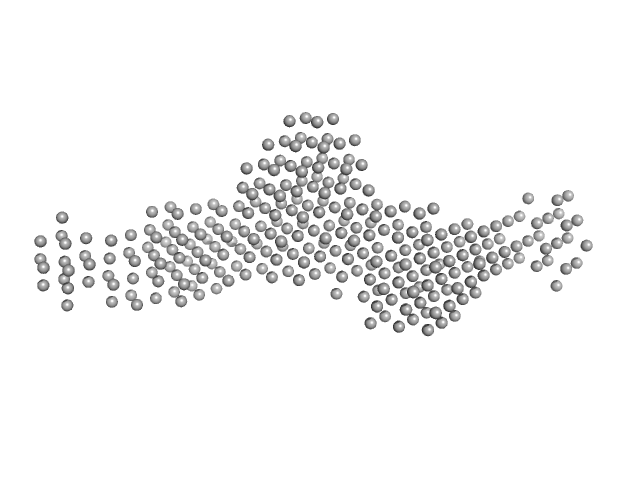
|
| Sample: |
SGT protein dimer, 92 kDa Leishmania braziliensis protein
|
| Buffer: |
20 mM Potassium Phosphate, 100 mM KCl, 10 mM EDTA, 1 mM B-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2014 May 14
|
Structural and functional studies of the Leishmania braziliensis SGT co-chaperone indicate that it shares structural features with HIP and can interact with both Hsp90 and Hsp70 with similar affinities.
Int J Biol Macromol 118(Pt A):693-706 (2018)
Coto ALS, Seraphim TV, Batista FAH, Dores-Silva PR, Barranco ABF, Teixeira FR, Gava LM, Borges JC
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.0 |
nm |
|
|
|
|
|
|
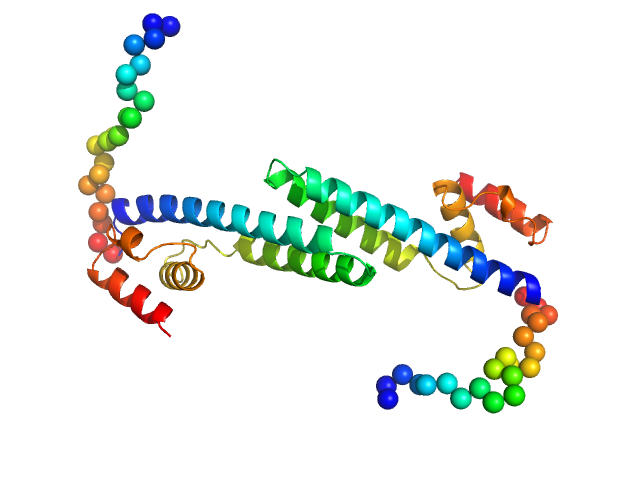
|
| Sample: |
Fluorescence recovery protein dimer, 28 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
2.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
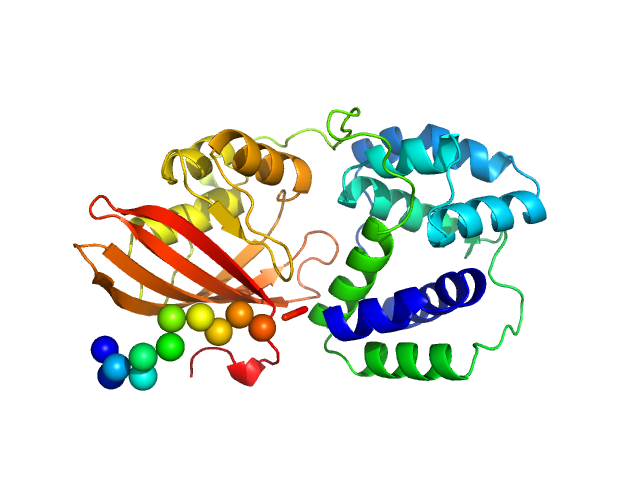
|
| Sample: |
Orange carotenoid-binding protein monomer, 34 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
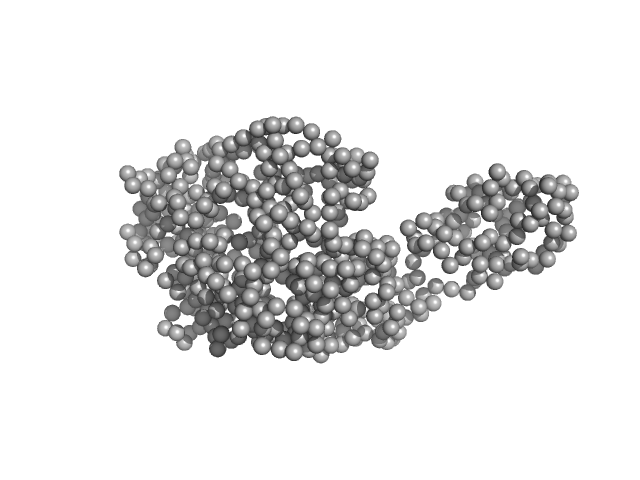
|
| Sample: |
Fluorescence recovery protein dimer, 28 kDa Synechocystis sp. PCC … protein
Orange carotenoid-binding protein monomer, 34 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
OCP-FRP protein complex topologies suggest a mechanism for controlling high light tolerance in cyanobacteria.
Nat Commun 9(1):3869 (2018)
Sluchanko NN, Slonimskiy YB, Shirshin EA, Moldenhauer M, Friedrich T, Maksimov EG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neural/ectodermal development factor IMP-L2 dimer, 60 kDa Drosophila melanogaster protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2011 Nov 20
|
Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun 9(1):3860 (2018)
Roed NK, Viola CM, Kristensen O, Schluckebier G, Norrman M, Sajid W, Wade JD, Andersen AS, Kristensen C, Ganderton TR, Turkenburg JP, De Meyts P, Brzozowski AM
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Insulin-like peptide 5 monomer, 5 kDa Drosophila melanogaster protein
Neural/ectodermal development factor IMP-L2 monomer, 30 kDa Drosophila melanogaster protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at ID14-3, ESRF on 2011 Nov 20
|
Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun 9(1):3860 (2018)
Roed NK, Viola CM, Kristensen O, Schluckebier G, Norrman M, Sajid W, Wade JD, Andersen AS, Kristensen C, Ganderton TR, Turkenburg JP, De Meyts P, Brzozowski AM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurexin 1a L5L6 monomer, 44 kDa protein
|
| Buffer: |
20 mM HEPES pH 8, 150 mM NaCl, 0.5mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, Sealy Center For Structural Biology, UTMB-G on 2016 Sep 6
|
Structural Plasticity of Neurexin 1α: Implications for its Role as Synaptic Organizer.
J Mol Biol 430(21):4325-4343 (2018)
Liu J, Misra A, Reddy MVVVS, White MA, Ren G, Rudenko G
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
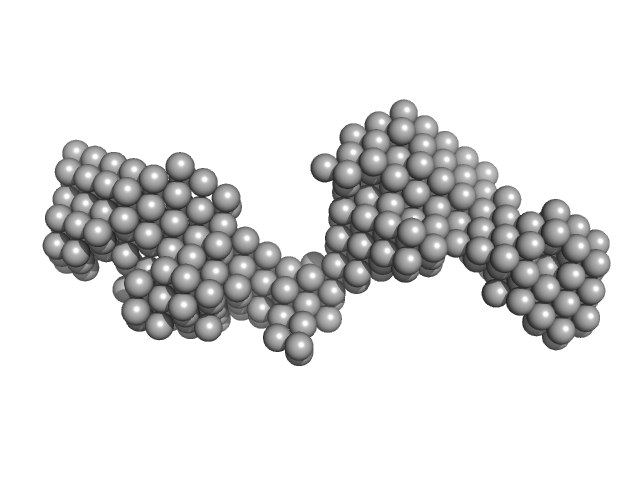
| Sample: |
Tegument protein UL37 monomer, 49 kDa Suid alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Inner tegument protein monomer, 62 kDa Human alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
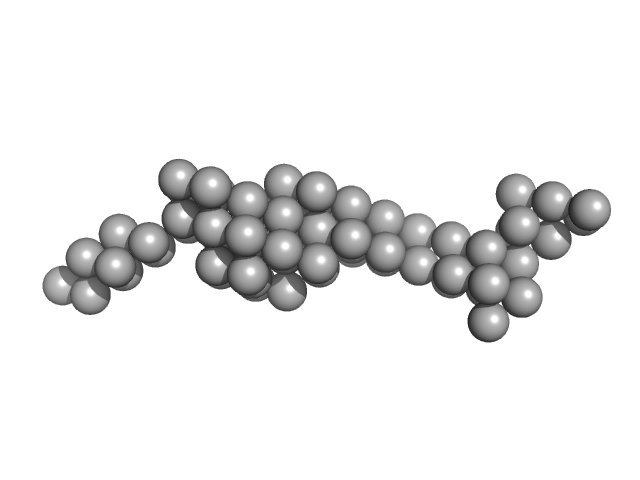
| Sample: |
Tegument protein UL37 monomer, 43 kDa Suid alphaherpesvirus 1 protein
|
| Buffer: |
100 mM HEPES 150 mM NaCl 5% glycerol 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), pH: 7.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Jun 3
|
The dynamic nature of the conserved tegument protein UL37 of herpesviruses.
J Biol Chem 293(41):15827-15839 (2018)
Koenigsberg AL, Heldwein EE
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
|
|