|
|
|
|
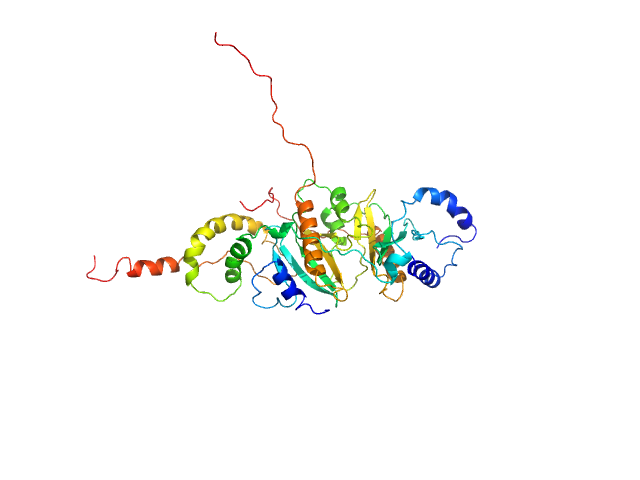
|
| Sample: |
SycH putative yopH targeting protein dimer, 32 kDa Yersinia pseudotuberculosis protein
Tyrosine-protein phosphatase YopH monomer, 14 kDa Yersinia pseudotuberculosis protein
|
| Buffer: |
50 mM HEPES 2mM TCEP, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Aug 1
|
Global Disordering in Stereo-Specific Protein Association
Biophysical Journal 112(3):33a (2017)
Gupta A, Reinartz I, Spilotros A, Jonna V, Hofer A, Svergun D, Schug A, Wolf-Watz M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-regulated outer membrane lipoprotein FrpD monomer, 27 kDa Neisseria meningitidis protein
|
| Buffer: |
10 mM Tris-HCl 150 mM NaCl 0.01% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Oct 19
|
Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep 7:40408 (2017)
Sviridova E, Rezacova P, Bondar A, Veverka V, Novak P, Schenk G, Svergun DI, Kuta Smatanova I, Bumba L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-regulated outer membrane lipoprotein FrpD monomer, 27 kDa Neisseria meningitidis protein
Iron-regulated protein FrpC monomer, 46 kDa Neisseria meningitidis protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl 0.01% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Oct 19
|
Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep 7:40408 (2017)
Sviridova E, Rezacova P, Bondar A, Veverka V, Novak P, Schenk G, Svergun DI, Kuta Smatanova I, Bumba L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
39 kDa FK506-binding nuclear protein tetramer, 163 kDa Drosophila melanogaster protein
|
| Buffer: |
50 mM Na2HPO4 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2014 Nov 14
|
Nucleoplasmin-like domain of FKBP39 from Drosophila melanogaster forms a tetramer with partly disordered tentacle-like C-terminal segments.
Sci Rep 7:40405 (2017)
Kozłowska M, Tarczewska A, Jakób M, Bystranowska D, Taube M, Kozak M, Czarnocki-Cieciura M, Dziembowski A, Orłowski M, Tkocz K, Ożyhar A
|
| RgGuinier |
5.7 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleasome Core Particle with Widom 601 DNA - HISTONE H2A-H2B Heterodimer dimer, 48 kDa Xenopus laevis protein
Nucleasome Core Particle with Widom 601 DNA - HISTONE H3-H4 Heterodimer dimer, 46 kDa Xenopus laevis protein
Nucleasome Core Particle with Widom 601 DNA - dsDNA monomer, 92 kDa Xenopus laevis DNA
|
| Buffer: |
20 mM Tris-Cl, 0.1 mM EDTA, 0.1 mM DTT, 50% sucrose, 1.2 M NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Apr 14
|
Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core.
Proc Natl Acad Sci U S A 114(2):334-339 (2017)
Chen Y, Tokuda JM, Topping T, Meisburger SP, Pabit SA, Gloss LM, Pollack L
|
|
|
|
|
|
|
|
| Sample: |
Nucleasome Core Particle with Widom 601 DNA - HISTONE H2A-H2B Heterodimer dimer, 48 kDa Xenopus laevis protein
Nucleasome Core Particle with Widom 601 DNA - HISTONE H3-H4 Heterodimer dimer, 46 kDa Xenopus laevis protein
Nucleasome Core Particle with Widom 601 DNA - dsDNA monomer, 92 kDa Xenopus laevis DNA
|
| Buffer: |
20 mM Tris-Cl, 0.1 mM EDTA, 0.1 mM DTT, 50% sucrose, 1.9 M NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2014 Apr 14
|
Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core.
Proc Natl Acad Sci U S A 114(2):334-339 (2017)
Chen Y, Tokuda JM, Topping T, Meisburger SP, Pabit SA, Gloss LM, Pollack L
|
|
|
|
|
|
|

|
| Sample: |
Outer membrane protein IcsA (53-758) monomer, 72 kDa Shigella flexneri protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 10 mM CaCl2 3% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 May 18
|
The Shigella Virulence Factor IcsA Relieves N-WASP Autoinhibition by Displacing the Verprolin Homology/Cofilin/Acidic (VCA) Domain.
J Biol Chem 292(1):134-145 (2017)
Mauricio RP, Jeffries CM, Svergun DI, Deane JE
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
S-layer protein monomer, 116 kDa Lysinibacillus sphaericus protein
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Analysis of self-assembly of S-layer protein slp-B53 from Lysinibacillus sphaericus.
Eur Biophys J 46(1):77-89 (2017)
Liu J, Falke S, Drobot B, Oberthuer D, Kikhney A, Guenther T, Fahmy K, Svergun D, Betzel C, Raff J
|
| RgGuinier |
5.8 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
495 |
nm3 |
|
|
|
|
|
|
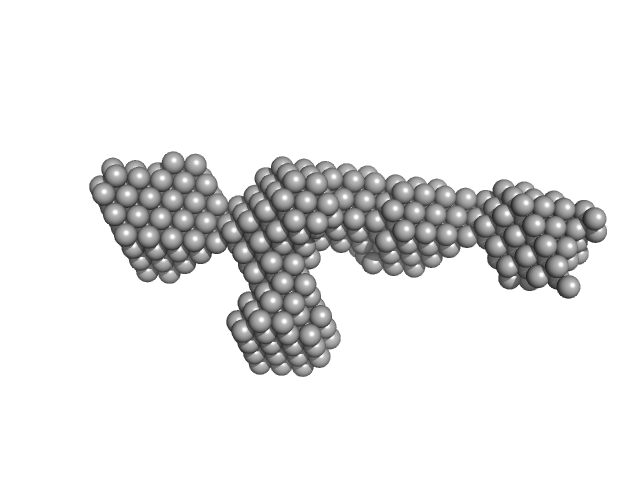
|
| Sample: |
S-layer protein monomer, 116 kDa Lysinibacillus sphaericus protein
|
| Buffer: |
Water with Ca2+, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Analysis of self-assembly of S-layer protein slp-B53 from Lysinibacillus sphaericus.
Eur Biophys J 46(1):77-89 (2017)
Liu J, Falke S, Drobot B, Oberthuer D, Kikhney A, Guenther T, Fahmy K, Svergun D, Betzel C, Raff J
|
| RgGuinier |
6.4 |
nm |
| Dmax |
28.1 |
nm |
| VolumePorod |
609 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
S-layer protein monomer, 116 kDa Lysinibacillus sphaericus protein
|
| Buffer: |
Water with Mg2+, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 2
|
Analysis of self-assembly of S-layer protein slp-B53 from Lysinibacillus sphaericus.
Eur Biophys J 46(1):77-89 (2017)
Liu J, Falke S, Drobot B, Oberthuer D, Kikhney A, Guenther T, Fahmy K, Svergun D, Betzel C, Raff J
|
| RgGuinier |
6.6 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
597 |
nm3 |
|
|