|
|
|
|
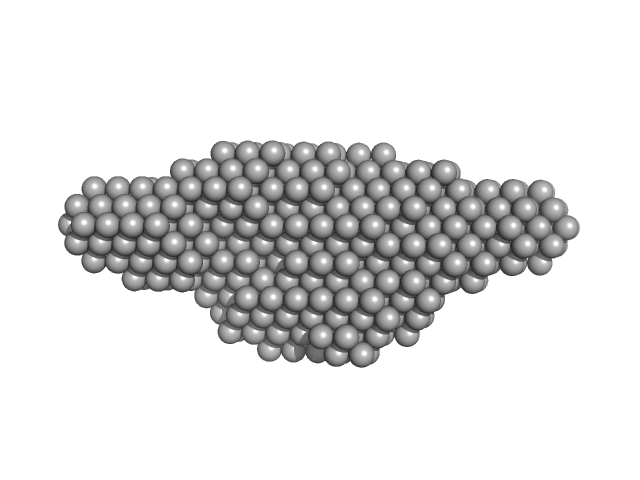
|
| Sample: |
Leishmania braziliensis heat shock protein 90 (Hsp90) dimer, 166 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
5.3 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
380 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Leishmania braziliensis heat shock protein 90 (Hsp90) N domain monomer, 26 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Leishmania braziliensis heat shock protein 90 (Hsp90) N and M domains monomer, 62 kDa Leishmania braziliensis protein
|
| Buffer: |
25 Tris mM 100 mM NaCl 1 mM 2-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2011 Sep 1
|
Insights on the structural dynamics of Leishmania braziliensis Hsp90 molecular chaperone by small angle X-ray scattering.
Int J Biol Macromol 97:503-512 (2017)
Seraphim TV, Silva KP, Dores-Silva PR, Barbosa LR, Borges JC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
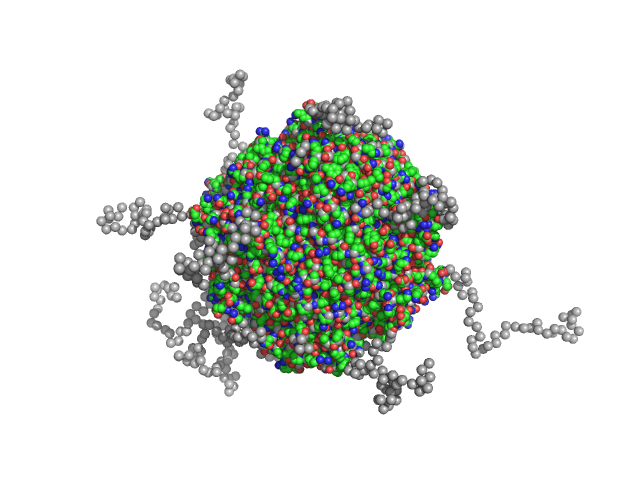
|
| Sample: |
N-terminal truncated DNA protection during starvation protein 2 dodecamer, 279 kDa Deinococcus radiodurans R1 protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
SAXS Structural Studies of Dps from Deinococcus radiodurans Highlights the Conformation of the Mobile N-Terminal Extensions.
J Mol Biol 429(5):667-687 (2017)
Santos SP, Cuypers MG, Round A, Finet S, Narayanan T, Mitchell EP, Romão CV
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
445 |
nm3 |
|
|
|
|
|
|
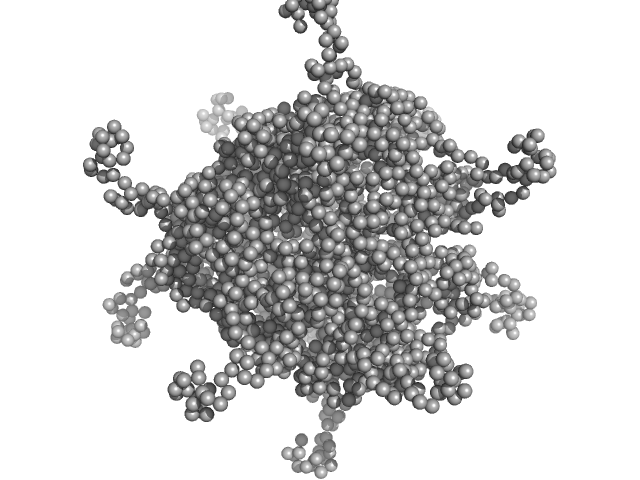
|
| Sample: |
DNA protection during starvation protein 1 dodecamer, 276 kDa Deinococcus radiodurans R1 protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
SAXS Structural Studies of Dps from Deinococcus radiodurans Highlights the Conformation of the Mobile N-Terminal Extensions.
J Mol Biol 429(5):667-687 (2017)
Santos SP, Cuypers MG, Round A, Finet S, Narayanan T, Mitchell EP, Romão CV
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
437 |
nm3 |
|
|
|
|
|
|
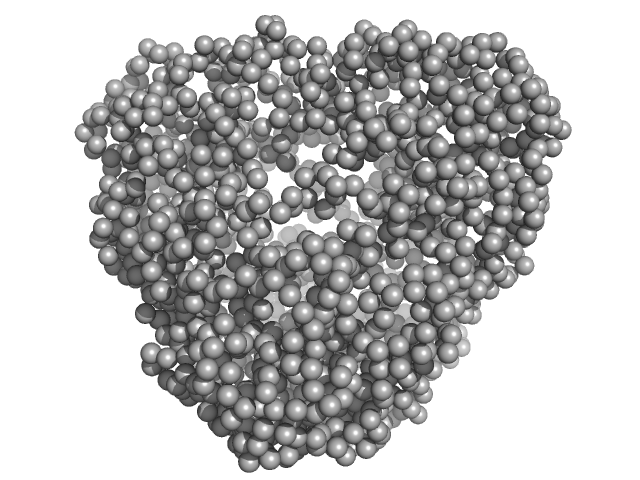
|
| Sample: |
N-terminal truncated DNA protection during starvation protein 1 dodecamer, 216 kDa Deinococcus radiodurans R1 protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
SAXS Structural Studies of Dps from Deinococcus radiodurans Highlights the Conformation of the Mobile N-Terminal Extensions.
J Mol Biol 429(5):667-687 (2017)
Santos SP, Cuypers MG, Round A, Finet S, Narayanan T, Mitchell EP, Romão CV
|
| RgGuinier |
3.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
291 |
nm3 |
|
|
|
|
|
|
|
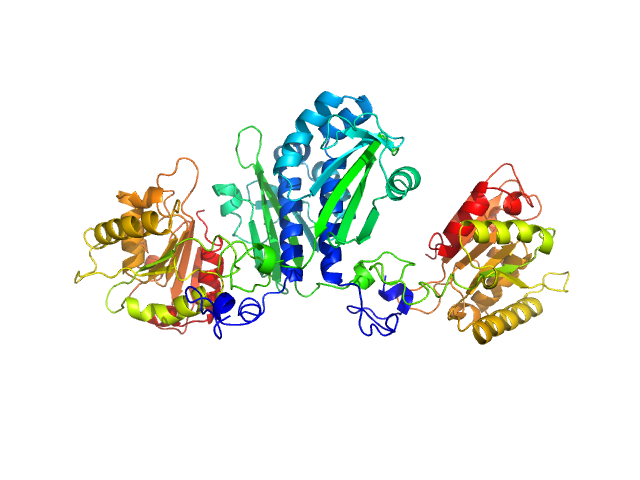
| Sample: |
Citrate-binding CitAP domain fused to lipase A of Bacillus subtilis BsLA dimer, 77 kDa protein
|
| Buffer: |
10 mM glycine buffer, 10 mM NaCl, 1 mM sodium citrate, pH: 10 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 6
|
A combination of mutational and computational scanning guides the design of an artificial ligand-binding controlled lipase.
Sci Rep 7:42592 (2017)
Kaschner M, Schillinger O, Fettweiss T, Nutschel C, Krause F, Fulton A, Strodel B, Stadler A, Jaeger KE, Krauss U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
Citrate-binding CitAP domain fused to lipase A of Bacillus subtilis BsLA dimer, 77 kDa protein
|
| Buffer: |
10 mM glycine buffer, 10 mM NaCl, pH: 10 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Feb 6
|
A combination of mutational and computational scanning guides the design of an artificial ligand-binding controlled lipase.
Sci Rep 7:42592 (2017)
Kaschner M, Schillinger O, Fettweiss T, Nutschel C, Krause F, Fulton A, Strodel B, Stadler A, Jaeger KE, Krauss U
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.8 |
nm |
|
|
|
|
|
|
|
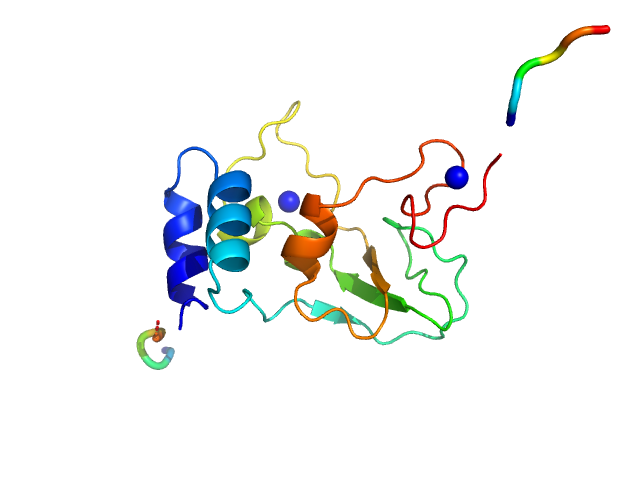
| Sample: |
HCoV-229E Non-structural protein 10 monomer, 15 kDa Human coronavirus 229E protein
|
| Buffer: |
25 mM HEPES 280 mM NaCl 2 mM DTT 500 µM ZnCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 25
|
Human alphacoronavirus non-structural protein Nsp10
Sven Falke,
Al Kikhney
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.8 |
nm |
|
|
|
|
|
|
|
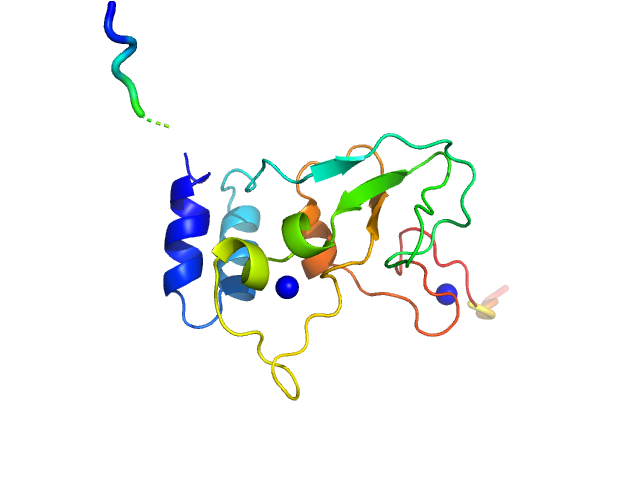
| Sample: |
HCoV-229E Non-structural protein 10 monomer, 15 kDa Human coronavirus 229E protein
|
| Buffer: |
25 mM HEPES 400 mM NaCl 1 mM EDTA 5% glycerol 40 mM NaH2PO4, pH: 7.9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2011 Sep 8
|
Human alphacoronavirus non-structural protein Nsp10
Sven Falke,
Al Kikhney
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.9 |
nm |
|
|