|
|
|
|
|
| Sample: |
AIR-3A 2'FU dimer, 13 kDa RNA
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AIR-3A G18U tetramer, 25 kDa RNA
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Functional binding region (187-385) of the pneumococcal serine-rich repeat protein monomer, 22 kDa Streptococcus pneumoniae protein
|
| Buffer: |
PBS 5 % Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Feb 27
|
The BR domain of PsrP interacts with extracellular DNA to promote bacterial aggregation; structural insights into pneumococcal biofilm formation.
Sci Rep 6:32371 (2016)
Schulte T, Mikaelsson C, Beaussart A, Kikhney A, Deshmukh M, Wolniak S, Pathak A, Ebel C, Löfling J, Fogolari F, Henriques-Normark B, Dufrêne YF, Svergun D, Nygren PÅ, Achour A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Functional binding region (120-395) of the pneumococcal serine-rich repeat protein monomer, 30 kDa Streptococcus pneumoniae protein
|
| Buffer: |
PBS 5 % Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Feb 27
|
The BR domain of PsrP interacts with extracellular DNA to promote bacterial aggregation; structural insights into pneumococcal biofilm formation.
Sci Rep 6:32371 (2016)
Schulte T, Mikaelsson C, Beaussart A, Kikhney A, Deshmukh M, Wolniak S, Pathak A, Ebel C, Löfling J, Fogolari F, Henriques-Normark B, Dufrêne YF, Svergun D, Nygren PÅ, Achour A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA 22mer monomer, 12 kDa DNA
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 5% glycerol 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 6
|
The flexibility of a homeodomain transcription factor heterodimer and its allosteric regulation by DNA binding.
FEBS J 283(16):3134-54 (2016)
Mathiasen L, Valentini E, Boivin S, Cattaneo A, Blasi F, Svergun DI, Bruckmann C
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aureochrome 1a bZIP-LOV module dimer, 57 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
50 mM Tris 50 mM boric acid 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aureochrome 1a bZIP-LOV module dimer, 57 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Aureochrome 1a bZIP-LOV module dimer, 57 kDa Phaeodactylum tricornutum protein
|
| Buffer: |
10 mM Tris 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Nov 6
|
Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes.
Nucleic Acids Res 44(12):5957-70 (2016)
Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen LO, Kottke T
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|

|
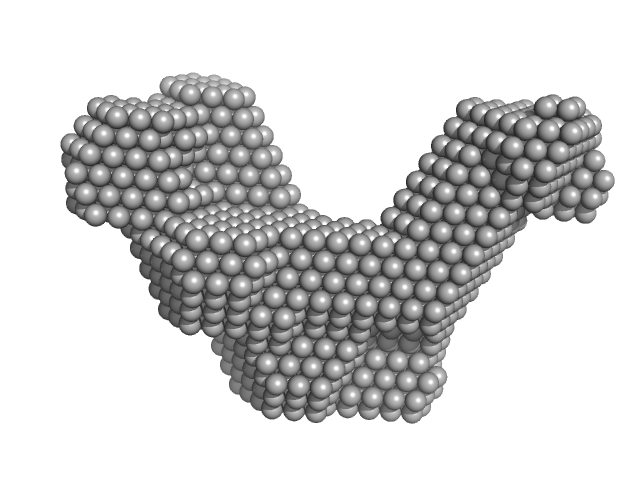
| Sample: |
Short-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 14 kDa Shewanella putrefaciens RNA
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Trimeric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) trimer, 112 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 27
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Cas6f: CRISPR/Cas Type I-F Cascade component (CRISPR-associated protein, Csy4 family) monomer, 21 kDa Shewanella putrefaciens protein
Cas5fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1822) monomer, 38 kDa Shewanella putrefaciens protein
Hexameric Cas7fv: CRISPR/Cas Type I-F Cascade component (Uncharacterized protein, Sputcn32_1821) hexamer, 223 kDa Shewanella putrefaciens protein
Wildtype-crRNA: CRISPR/Cas Type I-F Cascade component monomer, 19 kDa RNA
|
| Buffer: |
50 mM HEPES 150 mM NaCl 1mM DTT 1mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 27
|
Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Nucleic Acids Res 44(12):5872-82 (2016)
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L
|
| RgGuinier |
5.4 |
nm |
| Dmax |
18.4 |
nm |
|
|