|
|
|
|
|
| Sample: |
Netrin unc-6 monomer, 52 kDa Caenorhabditis elegans protein
Isoform a of Netrin receptor unc-5 monomer, 38 kDa Caenorhabditis elegans protein
Heparin, porcine intestinal mucosa monomer, 15 kDa Sus scrofa domesticus
Netrin receptor unc-40 monomer, 118 kDa Caenorhabditis elegans protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, 100 mM MgSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Dec 11
|
Structural insights into the formation of repulsive netrin guidance complexes.
Sci Adv 10(7):eadj8083 (2024)
Priest JM, Nichols EL, Smock RG, Hopkins JB, Mendoza JL, Meijers R, Shen K, Özkan E
|
| RgGuinier |
10.6 |
nm |
| Dmax |
36.5 |
nm |
| VolumePorod |
3410 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NHLP leader peptide family natural product (I56V) monomer, 9 kDa Methylovulum psychrotolerans protein
|
| Buffer: |
20 mM sodium phosphate (pH 7.5), 100 mM NaCl and 48 µM FAD, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Pennsylvania State University on 2023 Jul 28
|
Disordered regions in proteusin peptides guide post-translational modification by a flavin-dependent RiPP brominase.
Nat Commun 15(1):1265 (2024)
Nguyen NA, Vidya FNU, Yennawar NH, Wu H, McShan AC, Agarwal V
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
High mobility group protein B1 (D189E, E202D, E215D) monomer, 25 kDa Rattus norvegicus protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 11
|
The acidic intrinsically disordered region of the inflammatory mediator HMGB1 mediates fuzzy interactions with CXCL12.
Nat Commun 15(1):1201 (2024)
Mantonico MV, De Leo F, Quilici G, Colley LS, De Marchis F, Crippa M, Mezzapelle R, Schulte T, Zucchelli C, Pastorello C, Carmeno C, Caprioglio F, Ricagno S, Giachin G, Ghitti M, Bianchi ME, Musco G
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
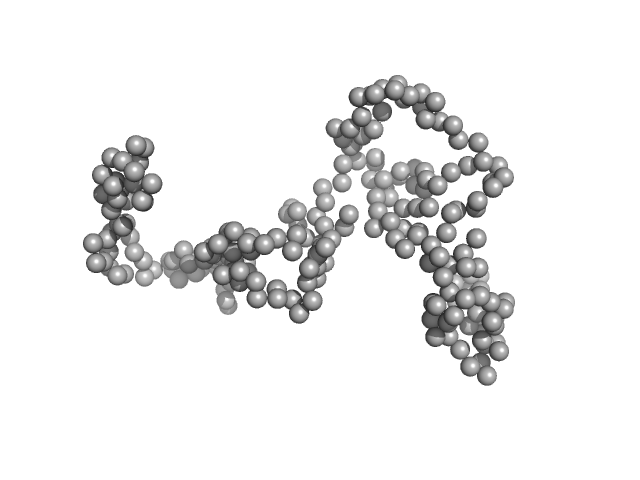
|
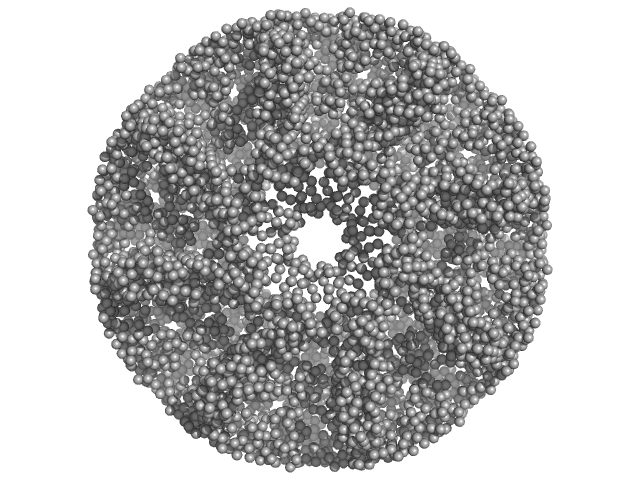
| Sample: |
DTMP kinase dimer, 57 kDa Brugia malayi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2018 Oct 7
|
Crystal Structure of the Brugia malayi Thymidylate Kinase-dTMP Complex and Small Angle X-ray Scattering Experiments Identifies Changes in the Dimeric Association Compared to the Human Homolog
Crystallography Reports 68(7):1150-1158 (2024)
Vishwakarma J, Sharma V, Kumar S, Ramachandran R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Carboxypeptidase-related protein 16-mer, 903 kDa Deinococcus radiodurans (strain … protein
|
| Buffer: |
20 mM Tris-Cl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Dec 25
|
Novel oligomeric assembly of S10-carboxypeptidase from Deinococcus radiodurans
Dr. Rahul Singh
|
| RgGuinier |
7.4 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
1180 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 27
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
705 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 27
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
704 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 27
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
702 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 29
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
699 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CH505TFchim.6R.SOSIP.664 Env glycoprotein trimer, 217 kDa HIV-1 group M protein
|
| Buffer: |
15 mM HEPES, 150 mM NaCl, pH: 7.1 |
| Experiment: |
SAXS
data collected at 14-ID-B (BioCARS), Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jul 30
|
Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv 10(5):eadj0396 (2024)
Bennett AL, Edwards R, Kosheleva I, Saunders C, Bililign Y, Williams A, Bubphamala P, Manosouri K, Anasti K, Saunders KO, Alam SM, Haynes BF, Acharya P, Henderson R
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
706 |
nm3 |
|
|