|
|
|
|
|
| Sample: |
Kinesin heavy chain (A515T) dimer, 221 kDa Drosophila melanogaster protein
|
| Buffer: |
25 mM HEPES/KOH, 150 mM NaCl, 1 mM MgCl2, 2 mM DTT, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Feb 19
|
Tropomyosin 1-I/C coordinates kinesin-1 and dynein motors during oskar mRNA transport.
Nat Struct Mol Biol (2024)
Heber S, McClintock MA, Simon B, Mehtab E, Lapouge K, Hennig J, Bullock SL, Ephrussi A
|
| RgGuinier |
10.0 |
nm |
| Dmax |
45.0 |
nm |
| VolumePorod |
920 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Kinesin heavy chain (A515T) dimer, 221 kDa Drosophila melanogaster protein
GH09289p dimer, 96 kDa Drosophila melanogaster protein
|
| Buffer: |
25 mM HEPES/KOH, 150 mM NaCl, 1 mM MgCl2, 2 mM DTT, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Feb 19
|
Tropomyosin 1-I/C coordinates kinesin-1 and dynein motors during oskar mRNA transport.
Nat Struct Mol Biol (2024)
Heber S, McClintock MA, Simon B, Mehtab E, Lapouge K, Hennig J, Bullock SL, Ephrussi A
|
| RgGuinier |
8.7 |
nm |
| Dmax |
45.0 |
nm |
| VolumePorod |
840 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein 1 monomer, 16 kDa Influenza B virus protein
5’ppp ds10 HP RNA monomer, 8 kDa RNA
|
| Buffer: |
40 mM ammonium acetate, pH 5.5, 225 mM NaCl, 5 mM CaCl2, 0.02% NaN3, 50 mM arginine, pH: 5.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Aug 6
|
The NS1 protein of influenza B virus binds 5’-triphosphorylated dsRNA to suppress RIG-I activation and the host antiviral response
(2024)
Woltz R, Schweibenz B, Tsutakawa S, Zhao C, Ma L, Shurina B, Hura G, John R, Vorobiev S, Swapna G, Solotchi M, Tainer J, Krug R, Patel S, Montelione G
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein 1 monomer, 16 kDa Influenza B virus protein
RNA top strand monomer, 5 kDa RNA
RNA bottom strand monomer, 5 kDa RNA
|
| Buffer: |
40 mM ammonium acetate, pH 5.5, 450 mM NaCl, 5 mM CaCl2, 0.02% NaN3, 50 mM arginine, pH: 5.5 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 May 18
|
The NS1 protein of influenza B virus binds 5’-triphosphorylated dsRNA to suppress RIG-I activation and the host antiviral response
(2024)
Woltz R, Schweibenz B, Tsutakawa S, Zhao C, Ma L, Shurina B, Hura G, John R, Vorobiev S, Swapna G, Solotchi M, Tainer J, Krug R, Patel S, Montelione G
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
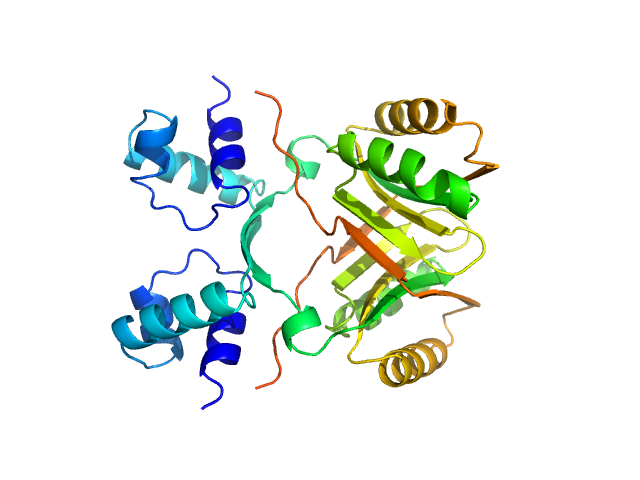
|
| Sample: |
AsnC family transcriptional regulator dimer, 36 kDa Haloferax mediterranei (strain … protein
|
| Buffer: |
50 mM Tris, 1.5 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 12
|
Global Lrp regulator protein from Haloferax mediterranei: Transcriptional analysis and structural characterization.
Int J Biol Macromol :129541 (2024)
Matarredona L, García-Bonete MJ, Guío J, Camacho M, Fillat MF, Esclapez J, Bonete MJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
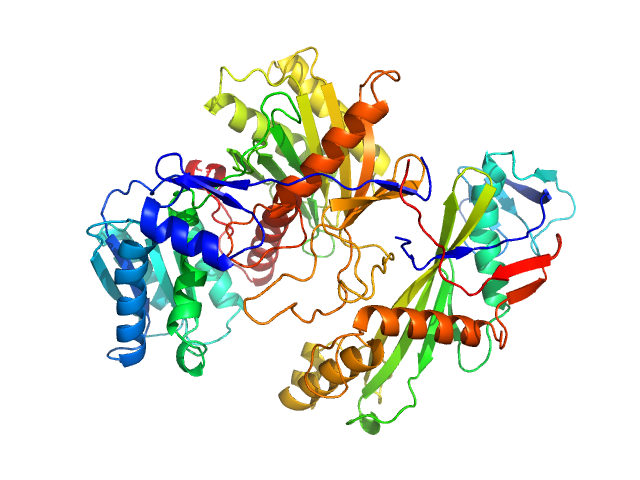
| Sample: |
Phosphocholinase AnkX monomer, 107 kDa Legionella pneumophila protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Oct 20
|
Legionella pneumophila effector AnkX
Wenhua Zhang
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
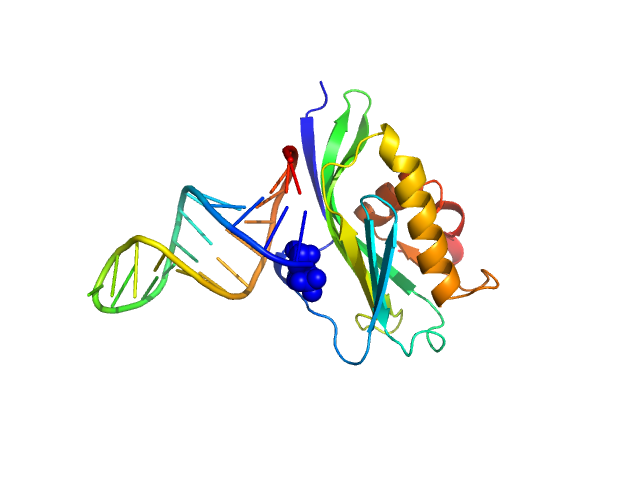
|
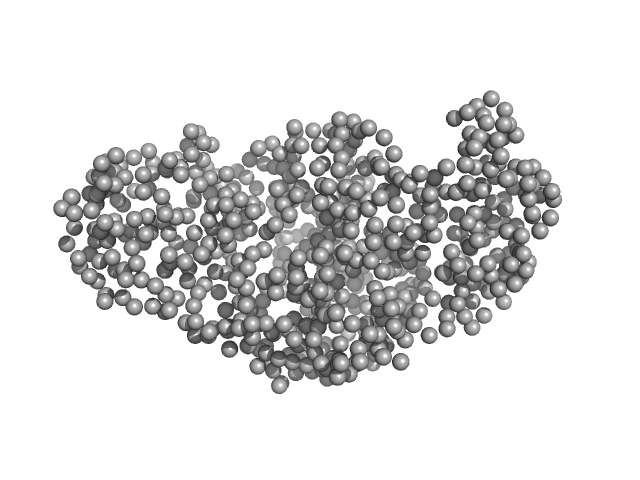
| Sample: |
Piwi protein AF_1318 (Archaeoglobus fulgidus AfAgo protein) monomer, 51 kDa Archaeoglobus fulgidus (strain … protein
Uncharacterized protein (AfAgo-N protein containing N-L1-L2 domains) monomer, 31 kDa Archaeoglobus fulgidus DSM … protein
5'-end phosphorylated DNA oligoduplex, 14 bp (MZ1288) monomer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH 7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
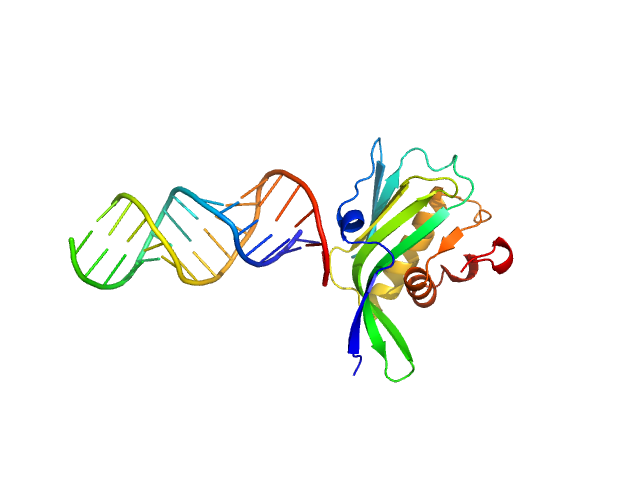
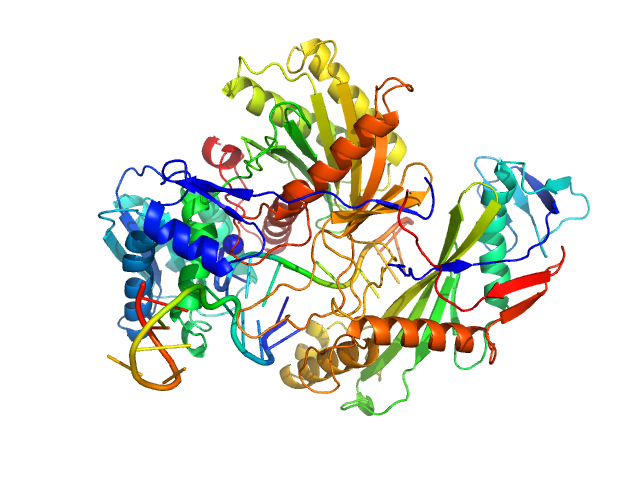
| Sample: |
Single-chain full Archaeoglobus fulgidus Argonaute monomer, 78 kDa Archaeoglobus fulgidus protein
5'-end phosphorylated DNA oligoduplex, 14 bp (MZ1288) monomer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
131 |
nm3 |
|
|
|
|
|
|
|
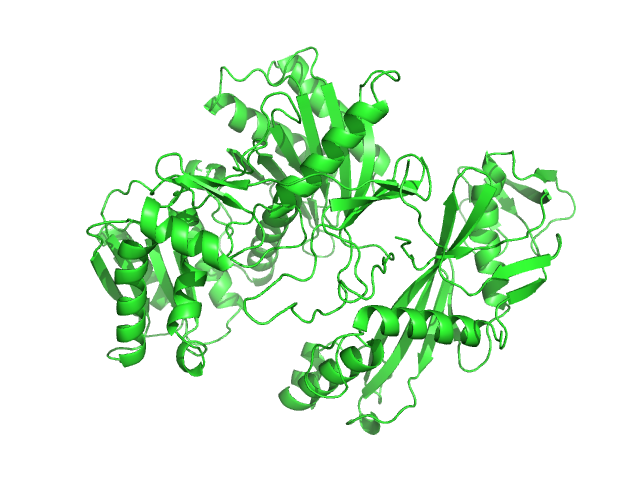
| Sample: |
Single-chain full Archaeoglobus fulgidus Argonaute monomer, 78 kDa Archaeoglobus fulgidus protein
5'-end phosphorylated DNA guide strand, 11 nt (MZ864) monomer, 3 kDa DNA
DNA target strand, 11 nt (MZ865) monomer, 3 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Aug 31
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Single-chain full Archaeoglobus fulgidus Argonaute variant monomer, 78 kDa Archaeoglobus fulgidus protein
|
| Buffer: |
20 mM TrisHCl pH 7.5, 500 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Aug 31
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
142 |
nm3 |
|
|