|
|
|
|
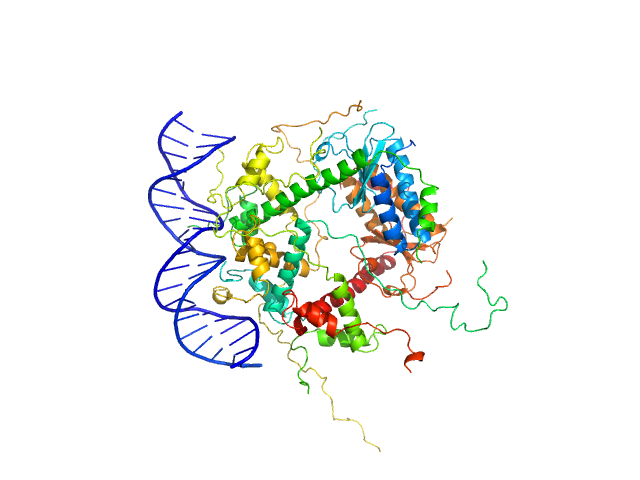
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
21-bp DNA operator fragment monomer, 13 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
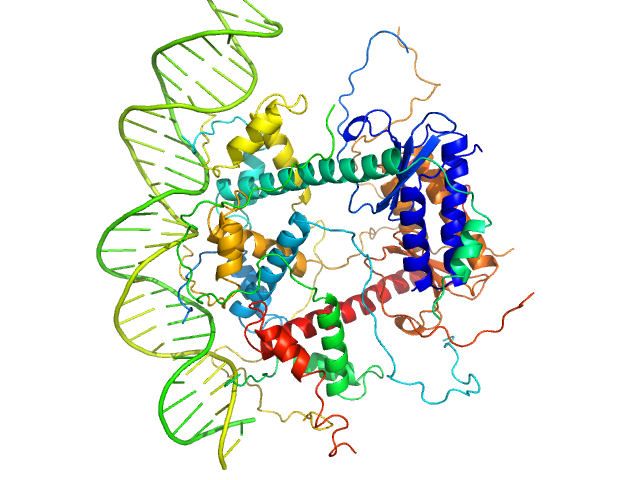
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
31-bp DNA operator box monomer, 19 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
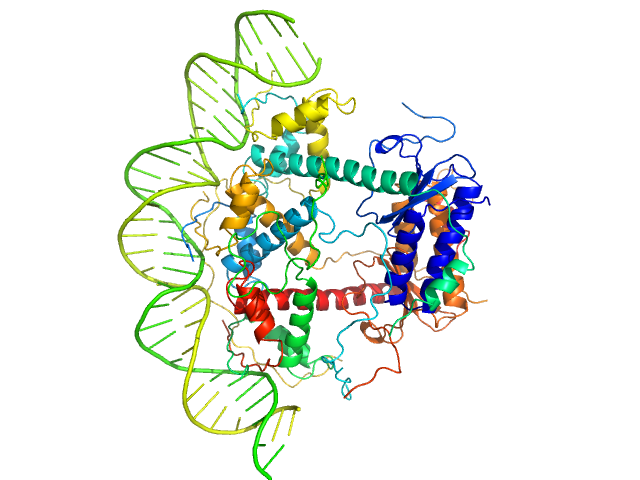
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
33-bp DNA operator fragment monomer, 20 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
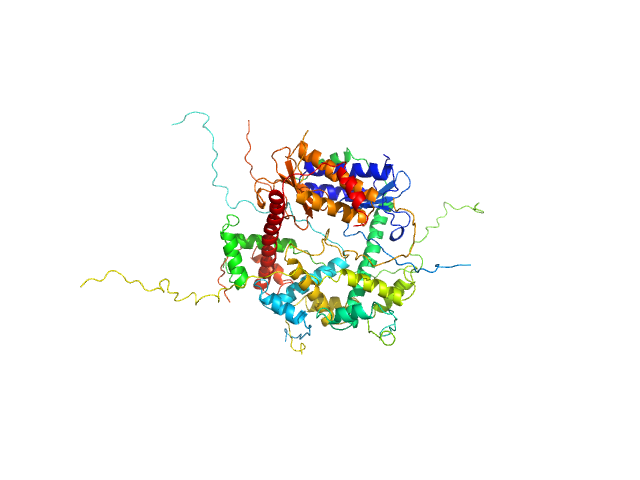
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 6
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
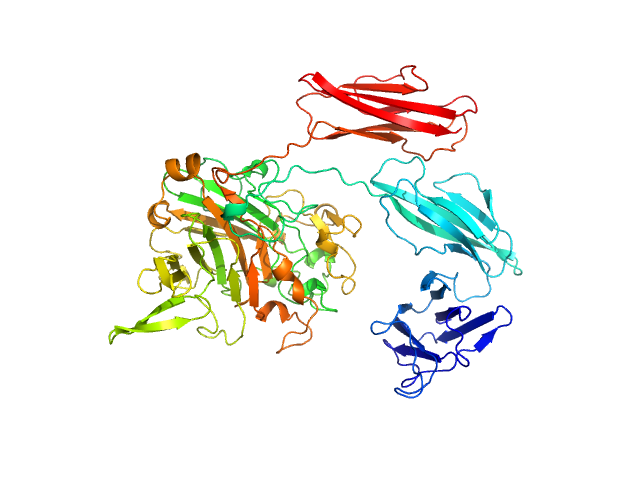
|
| Sample: |
Dockerin type I repeat monomer, 69 kDa Ruminococcus bromii protein
|
| Buffer: |
phosphate buffered saline, 1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
The Ruminococcus bromii amylosome protein Sas6 binds single and double helical α-glucan structures in starch.
Nat Struct Mol Biol (2024)
Photenhauer AL, Villafuerte-Vega RC, Cerqueira FM, Armbruster KM, Mareček F, Chen T, Wawrzak Z, Hopkins JB, Vander Kooi CW, Janeček Š, Ruotolo BT, Koropatkin NM
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Biofilm regulatory protein monomer, 34 kDa Streptococcus dysgalactiae subsp. … protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 1
|
Structural insights of an LCP protein-LytR-from Streptococcus dysgalactiae subs. dysgalactiae through biophysical and in silico methods.
Front Chem 12:1379914 (2024)
Paquete-Ferreira J, Freire F, Fernandes HS, Muthukumaran J, Ramos J, Bryton J, Panjkovich A, Svergun D, Santos MFA, Correia MAS, Fernandes AR, Romão MJ, Sousa SF, Santos-Silva T
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Biofilm regulatory protein monomer, 34 kDa Streptococcus dysgalactiae subsp. … protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 1
|
Structural insights of an LCP protein-LytR-from Streptococcus dysgalactiae subs. dysgalactiae through biophysical and in silico methods.
Front Chem 12:1379914 (2024)
Paquete-Ferreira J, Freire F, Fernandes HS, Muthukumaran J, Ramos J, Bryton J, Panjkovich A, Svergun D, Santos MFA, Correia MAS, Fernandes AR, Romão MJ, Sousa SF, Santos-Silva T
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
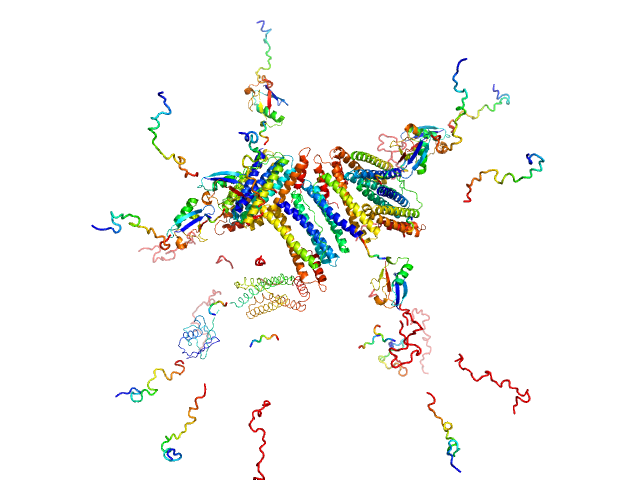
|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion) 24-mer, 755 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Jul 30
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
| RgGuinier |
7.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
2010 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion) octamer, 252 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Jul 17
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
618 |
nm3 |
|
|
|
|
|
|
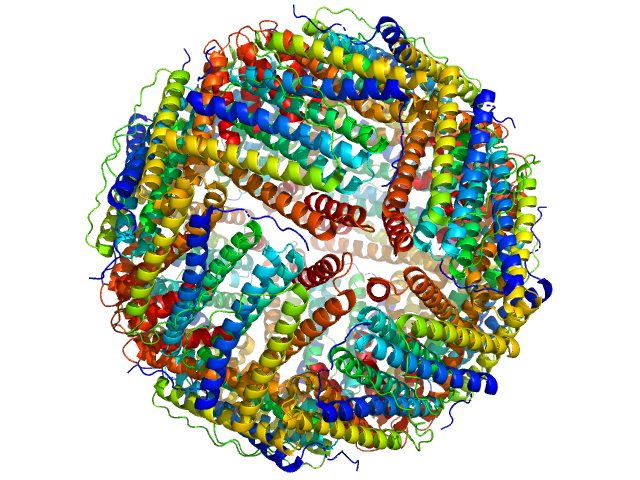
|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V) 24-mer, 480 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Dec 14
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
| RgGuinier |
5.6 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
674 |
nm3 |
|
|