|
|
|
|
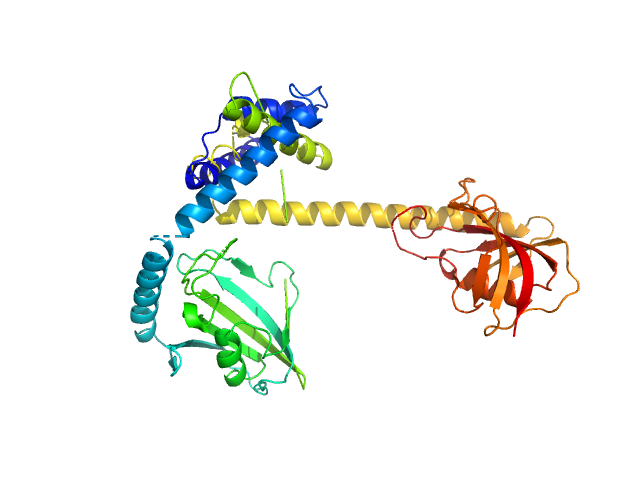
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
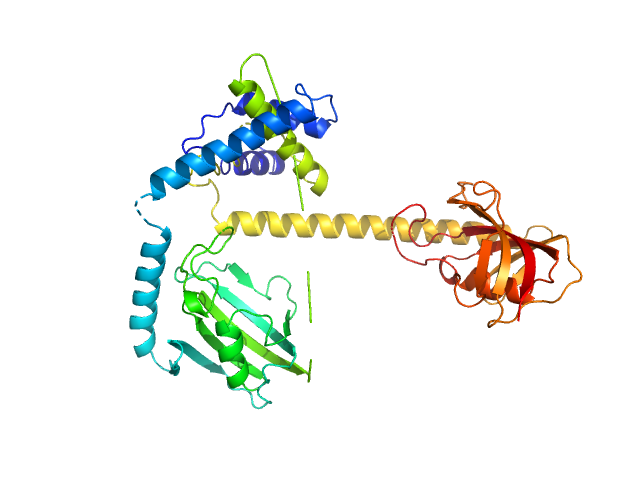
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
|
|
|
|
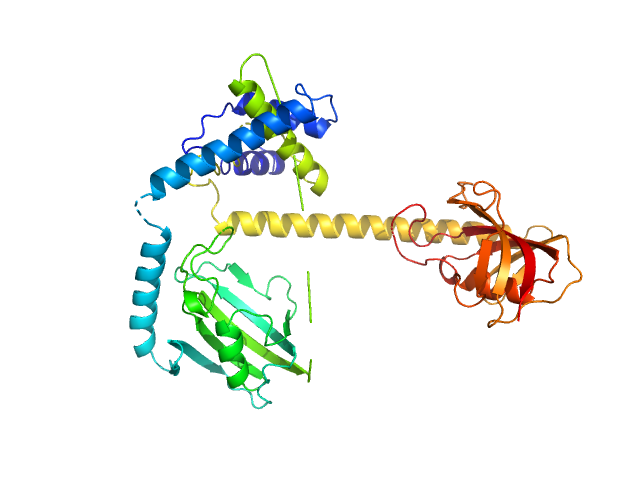
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 14 mer monomer, 5 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
Conformation and structural dynamics of the Xist lncRNA A-repeats
(2023)
Jones A, Gabel F, Bohn S, Wolfe G, Sattler M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 24 mer dimer, 16 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
Conformation and structural dynamics of the Xist lncRNA A-repeats
(2023)
Jones A, Gabel F, Bohn S, Wolfe G, Sattler M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 1 repeat dimer, 41 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
Conformation and structural dynamics of the Xist lncRNA A-repeats
(2023)
Jones A, Gabel F, Bohn S, Wolfe G, Sattler M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 2 repeats monomer, 34 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
Conformation and structural dynamics of the Xist lncRNA A-repeats
(2023)
Jones A, Gabel F, Bohn S, Wolfe G, Sattler M
|
| RgGuinier |
5.0 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 3 repeats monomer, 48 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
Conformation and structural dynamics of the Xist lncRNA A-repeats
(2023)
Jones A, Gabel F, Bohn S, Wolfe G, Sattler M
|
| RgGuinier |
6.3 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
97 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 4 repeats monomer, 62 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
Conformation and structural dynamics of the Xist lncRNA A-repeats
(2023)
Jones A, Gabel F, Bohn S, Wolfe G, Sattler M
|
| RgGuinier |
7.2 |
nm |
| Dmax |
34.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 5 repeats monomer, 81 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
Conformation and structural dynamics of the Xist lncRNA A-repeats
(2023)
Jones A, Gabel F, Bohn S, Wolfe G, Sattler M
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|