|
|
|
|
|
| Sample: |
Xist A-repeat lncRNA 6 repeats monomer, 96 kDa RNA
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 20
|
Conformation and structural dynamics of the Xist lncRNA A-repeats
(2023)
Jones A, Gabel F, Bohn S, Wolfe G, Sattler M
|
| RgGuinier |
12.9 |
nm |
| Dmax |
38.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Segment S(66-81) of the Neurofilament low intrinsically disordered tail domain monomer, 2 kDa house mouse protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Apr 20
|
Intramolecular structural heterogeneity altered by long-range contacts in an intrinsically disordered protein.
Proc Natl Acad Sci U S A 120(30):e2220180120 (2023)
Koren G, Meir S, Holschuh L, Mertens HDT, Ehm T, Yahalom N, Golombek A, Schwartz T, Svergun DI, Saleh OA, Dzubiella J, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Segment S(66-81) of the Neurofilament low intrinsically disordered tail domain monomer, 2 kDa house mouse protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Sep 15
|
Intramolecular structural heterogeneity altered by long-range contacts in an intrinsically disordered protein.
Proc Natl Acad Sci U S A 120(30):e2220180120 (2023)
Koren G, Meir S, Holschuh L, Mertens HDT, Ehm T, Yahalom N, Golombek A, Schwartz T, Svergun DI, Saleh OA, Dzubiella J, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Segment S(66-81) of the Neurofilament low intrinsically disordered tail domain monomer, 2 kDa house mouse protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Sep 15
|
Intramolecular structural heterogeneity altered by long-range contacts in an intrinsically disordered protein.
Proc Natl Acad Sci U S A 120(30):e2220180120 (2023)
Koren G, Meir S, Holschuh L, Mertens HDT, Ehm T, Yahalom N, Golombek A, Schwartz T, Svergun DI, Saleh OA, Dzubiella J, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Segment S(66-81) of the Neurofilament low intrinsically disordered tail domain monomer, 2 kDa house mouse protein
|
| Buffer: |
20 mM Tris, 500 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Sep 15
|
Intramolecular structural heterogeneity altered by long-range contacts in an intrinsically disordered protein.
Proc Natl Acad Sci U S A 120(30):e2220180120 (2023)
Koren G, Meir S, Holschuh L, Mertens HDT, Ehm T, Yahalom N, Golombek A, Schwartz T, Svergun DI, Saleh OA, Dzubiella J, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Segment S(66-81) of the Neurofilament low intrinsically disordered tail domain monomer, 2 kDa house mouse protein
|
| Buffer: |
20 mM Tris, 1 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Sep 15
|
Intramolecular structural heterogeneity altered by long-range contacts in an intrinsically disordered protein.
Proc Natl Acad Sci U S A 120(30):e2220180120 (2023)
Koren G, Meir S, Holschuh L, Mertens HDT, Ehm T, Yahalom N, Golombek A, Schwartz T, Svergun DI, Saleh OA, Dzubiella J, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Segment S(66-81) of the Neurofilament low intrinsically disordered tail domain monomer, 2 kDa house mouse protein
|
| Buffer: |
20 mM Tris, 3 M GdnHCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Sep 15
|
Intramolecular structural heterogeneity altered by long-range contacts in an intrinsically disordered protein.
Proc Natl Acad Sci U S A 120(30):e2220180120 (2023)
Koren G, Meir S, Holschuh L, Mertens HDT, Ehm T, Yahalom N, Golombek A, Schwartz T, Svergun DI, Saleh OA, Dzubiella J, Beck R
|
|
|
|
|
|
|
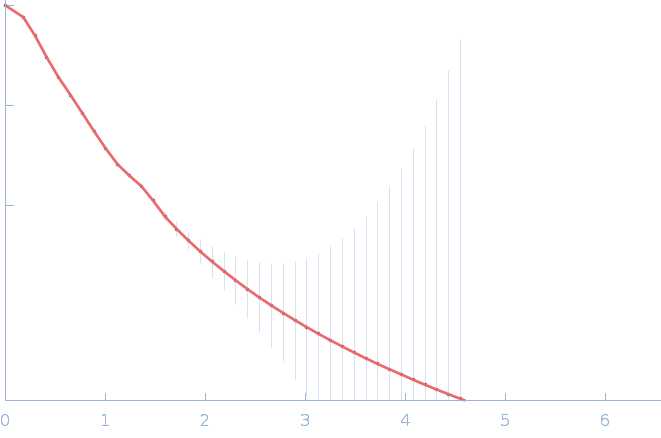
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
233 |
nm3 |
|
|
|
|
|
|
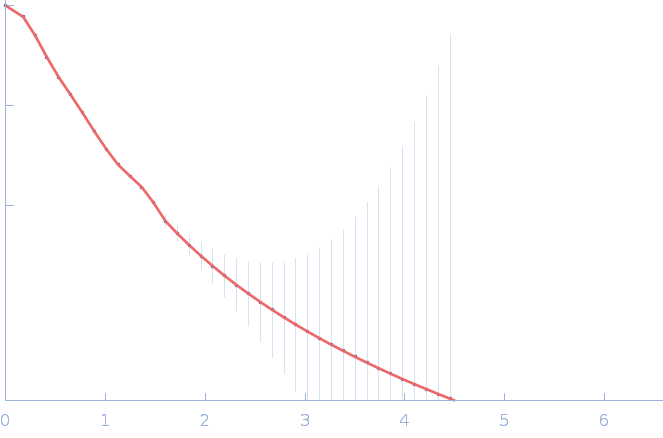
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
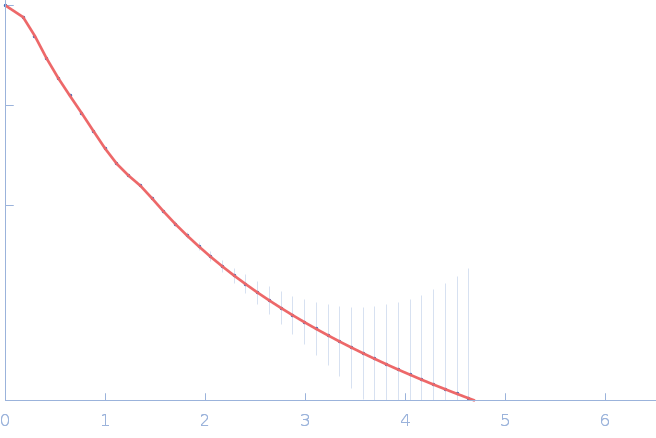
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
243 |
nm3 |
|
|