|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRT9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan higher order esters fraction (F4) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 0, 42 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 29
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
8.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
WGS project CABT00000000 data, contig 2.12 tetramer, 155 kDa Sordaria macrospora (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
9.6 |
nm |
| Dmax |
38.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Protein PAIR1 tetramer, 132 kDa Zea mays protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
8.0 |
nm |
| Dmax |
31.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
Meiotic recombination protein rec15 tetramer, 128 kDa Schizosaccharomyces pombe (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
6.9 |
nm |
| Dmax |
26.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
DUF2285 domain-containing protein monomer, 11 kDa Mesorhizobium loti protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 1
|
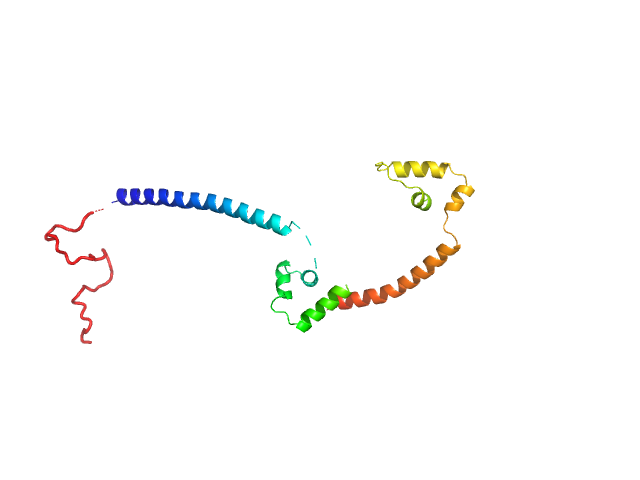
DUF2285 is a novel helix-turn-helix domain variant that orchestrates both activation and antiactivation of conjugative element transfer in proteobacteria.
Nucleic Acids Res (2023)
Jowsey WJ, Morris CRP, Hall DA, Sullivan JT, Fagerlund RD, Eto KY, Solomon PD, Mackay JP, Bond CS, Ramsay JP, Ronson CW
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
|
| Buffer: |
50mM Tris, 750mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Oct 10
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
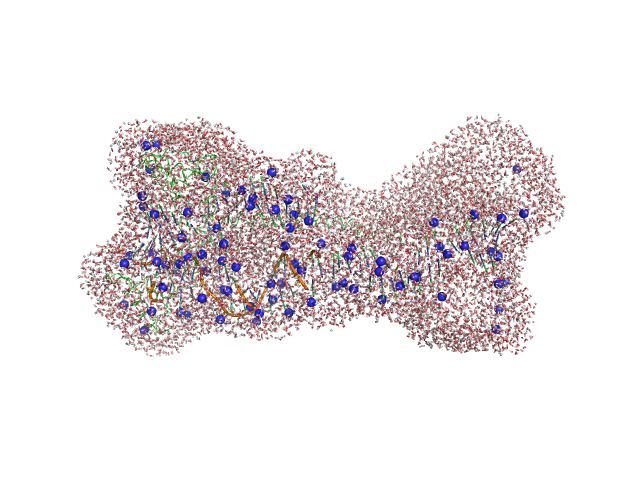
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
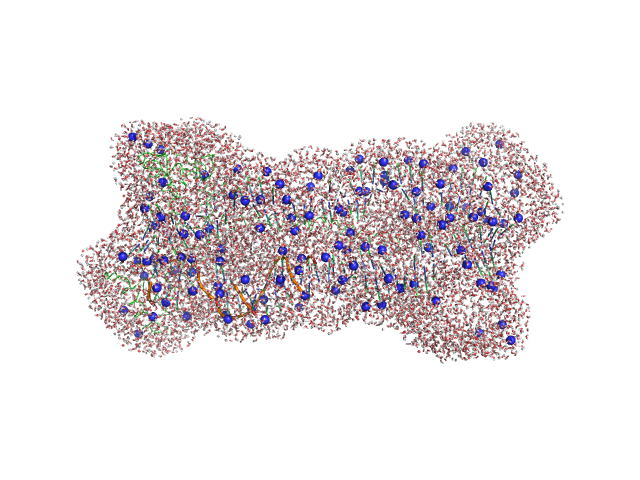
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
133 |
nm3 |
|
|
|
|
|
|
|
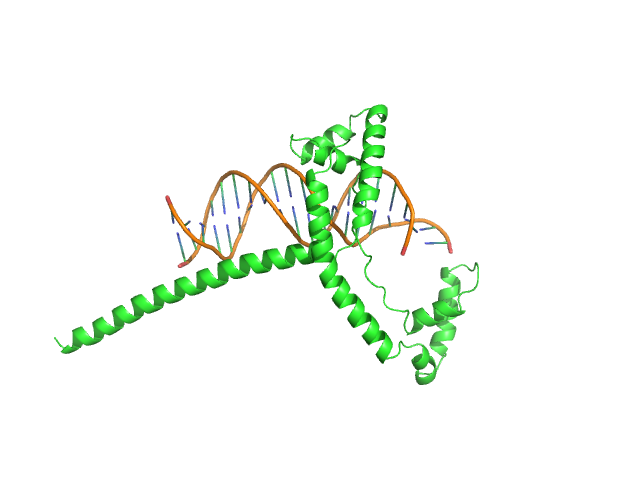
| Sample: |
Af2_20 DNA monomer, 12 kDa DNA
Gcf1p(Δ58) monomer, 22 kDa Candida albicans (strain … protein
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Monekypox DNA sequence 1 monomer, 6 kDa Monkeypox virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, and 0.2 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 23
|
Mapping and characterization of G‐quadruplexes in monkeypox genomes
Journal of Medical Virology 95(5) (2023)
Pereira H, Gemmill D, Siddiqui M, Vasudeva G, Patel T
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.4 |
nm |
| VolumePorod |
14 |
nm3 |
|
|