|
|
|
|
|
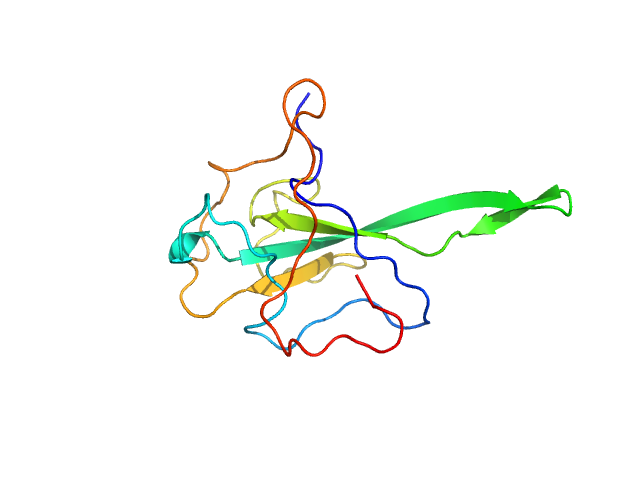
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 7 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRN9_fit1_model1.png)
|
| Sample: |
Polysorbate 20 (PS20) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 0, 45 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 2
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.6 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRP9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of Polysorbate 20 (PS20) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 0, 45 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 2
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
8.8 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRQ9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan monolaurate fraction (F2) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 20-mer, 24 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRR9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan monolaurate fraction (F2) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 22-mer, 26 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRS9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan higher order esters fraction (F4) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 0, 42 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 29
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.3 |
nm |
|
|