|
|
|
|
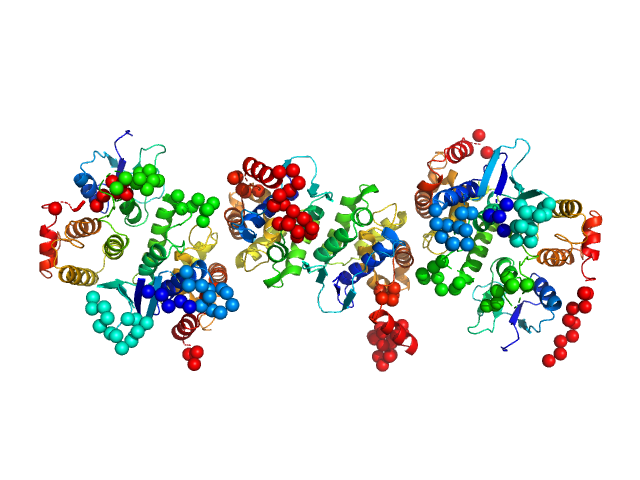
|
| Sample: |
Glutamine--tRNA ligase dimer, 46 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
TRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
|
|
|
|
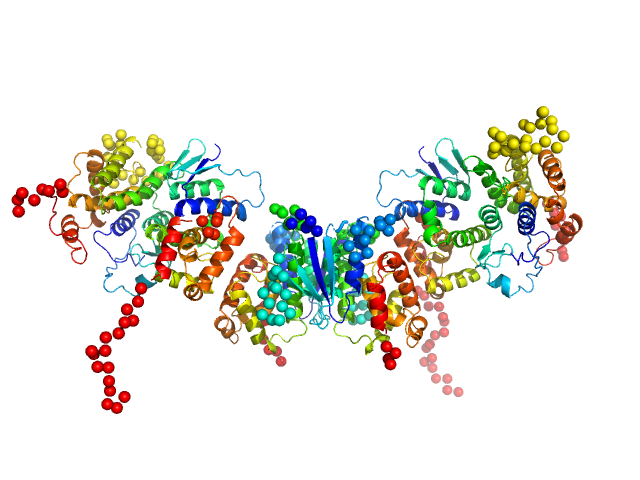
|
| Sample: |
Methionine--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
TRNA import protein tRIP dimer, 48 kDa Plasmodium berghei (strain … protein
Glutamate--tRNA ligase dimer, 55 kDa Plasmodium berghei (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 300 mM NaCl, 5% (v/v) glycerol, 0.005% (m/v) n-dodecyl beta-D-maltoside, 5 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 9
|
Solution x-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci :e4564 (2023)
Jaramillo Ponce JR, Théobald-Dietrich A, Bénas P, Paulus C, Sauter C, Frugier M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dihydroneopterin aldolase tetramer, 55 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5 and 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Apr 5
|
Structure of Helicobacter pylori dihydroneopterin aldolase suggests a fragment-based strategy for isozyme-specific inhibitor design.
Curr Res Struct Biol 5:100095 (2023)
Shaw GX, Fan L, Cherry S, Shi G, Tropea JE, Ji X
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
100 mM HEPES pH 7.5, 20 %(v/v) jeffamine M-600, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
The Relationship of Precursor Cluster Concentration in a Saturated Crystallization Solution to Long-Range Order During the Transition to the Solid Phase.
Acta Naturae 15(1):58-68 (2023)
Marchenkova MA, Boikova AS, Ilina KB, Konarev PV, Pisarevsky YV, Dyakova YA, Kovalchuk MV
|
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
100 mM sodium acetate, pH 4.6, 2.0 M sodium formate, pH: 4.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
The Relationship of Precursor Cluster Concentration in a Saturated Crystallization Solution to Long-Range Order During the Transition to the Solid Phase.
Acta Naturae 15(1):58-68 (2023)
Marchenkova MA, Boikova AS, Ilina KB, Konarev PV, Pisarevsky YV, Dyakova YA, Kovalchuk MV
|
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
200 mM K/Na tartrate, 100 mM tri-sodium citrate pH 5.6, 2.0 M ammonium sulfate, pH: 5.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
The Relationship of Precursor Cluster Concentration in a Saturated Crystallization Solution to Long-Range Order During the Transition to the Solid Phase.
Acta Naturae 15(1):58-68 (2023)
Marchenkova MA, Boikova AS, Ilina KB, Konarev PV, Pisarevsky YV, Dyakova YA, Kovalchuk MV
|
|
|
|
|
|
|
|
| Sample: |
Non-structural protein V monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
3.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein V (Y211A, Y212A, Y213A mutant) monomer, 15 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein V monomer, 9 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein V (ΔC-terminal and Y111A, Y112A, Y113A mutant) monomer, 9 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|