|
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
|
|
|
|
|
|
|
| Sample: |
Early E1A protein monomer, 13 kDa Human adenovirus C … protein
|
| Buffer: |
20 mM sodium phosphate pH 7.0, 200 mM NaCl, 1mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 24
|
Conformational buffering underlies functional selection in intrinsically disordered protein regions.
Nat Struct Mol Biol (2022)
González-Foutel NS, Glavina J, Borcherds WM, Safranchik M, Barrera-Vilarmau S, Sagar A, Estaña A, Barozet A, Garrone NA, Fernandez-Ballester G, Blanes-Mira C, Sánchez IE, de Prat-Gay G, Cortés J, Bernadó P, Pappu RV, Holehouse AS, Daughdrill GW, Chemes LB
|
| RgGuinier |
3.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Rhoptry kinase family protein tetramer, 143 kDa Toxoplasma gondii (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2016 Apr 19
|
Divergent kinase WNG1 is regulated by phosphorylation of an atypical activation sub-domain.
Biochem J (2022)
Dewangan PS, Beraki TG, Paiz EA, Appiah Mensah D, Chen Z, Reese ML
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Lysin [Streptococcus phage P7951] hexamer, 65 kDa Streptococcus phage P7951 protein
|
| Buffer: |
50 mM HEPES, 500 mM NaCl, and 1% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 21
|
On the Occurrence and Multimerization of Two-Polypeptide Phage Endolysins Encoded in Single Genes.
Microbiol Spectr :e0103722 (2022)
Pinto D, Gonçalo R, Louro M, Silva MS, Hernandez G, Cordeiro TN, Cordeiro C, São-José C
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit MurT monomer, 52 kDa Streptococcus pneumoniae (strain … protein
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit GatD monomer, 29 kDa Streptococcus pneumoniae (strain … protein
|
| Buffer: |
50 mM Hepes, 10 mM MgCl2, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 23
|
Unravelling the reaction mechanism of glutamate amidation in Staphylococcus aureus peptidoglycan
PhD thesis, NOVA University Lisbon - (2022)
Francisco Miguel Piçarra Leisico, Mertens HD
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit MurT monomer, 49 kDa Staphylococcus aureus (strain … protein
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit GatD monomer, 28 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
100 mM Tris-HCl, 500 mM NaCl, 10 mM MgCl2, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 29
|
Unravelling the reaction mechanism of glutamate amidation in Staphylococcus aureus peptidoglycan
PhD thesis, NOVA University Lisbon - (2022)
Francisco Miguel Piçarra Leisico, Mertens HD
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
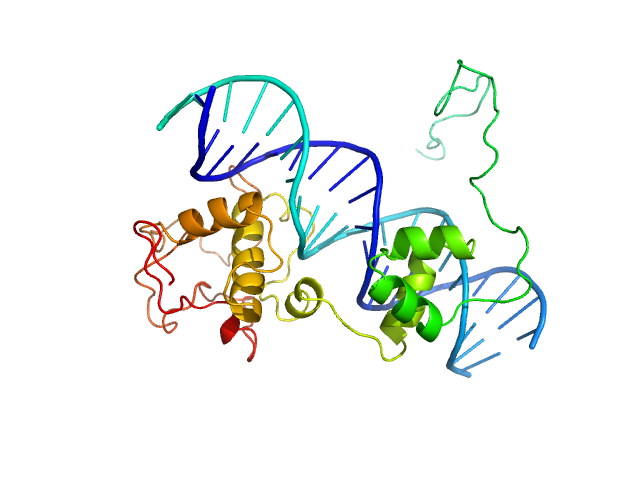
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
21mer dsDNA monomer, 13 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
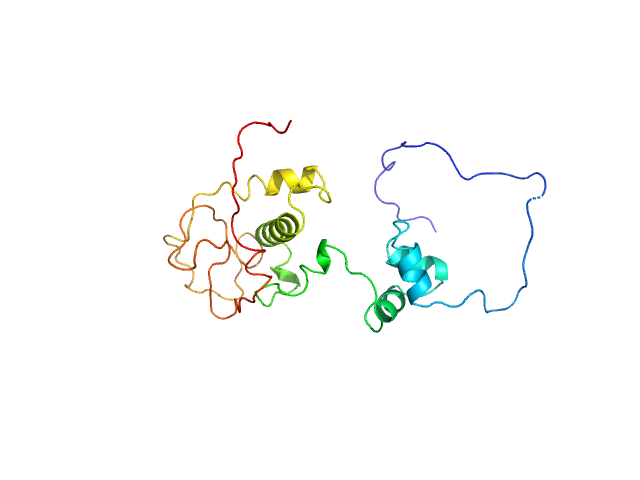
|
| Sample: |
Immunity repressor monomer, 24 kDa Mycobacterium phage TipsytheTRex protein
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
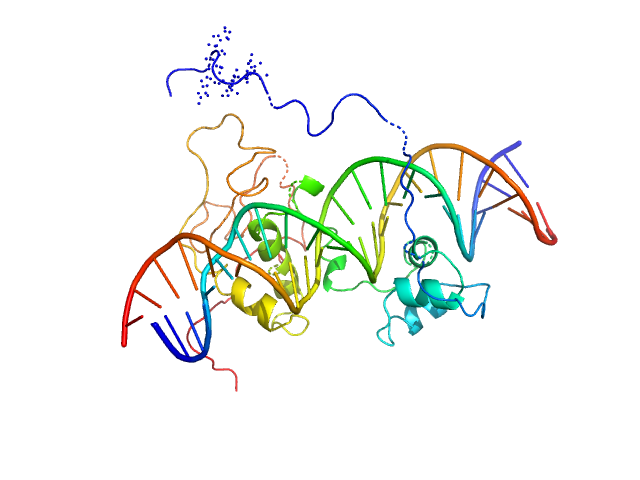
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
24mer dsDNA monomer, 15 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
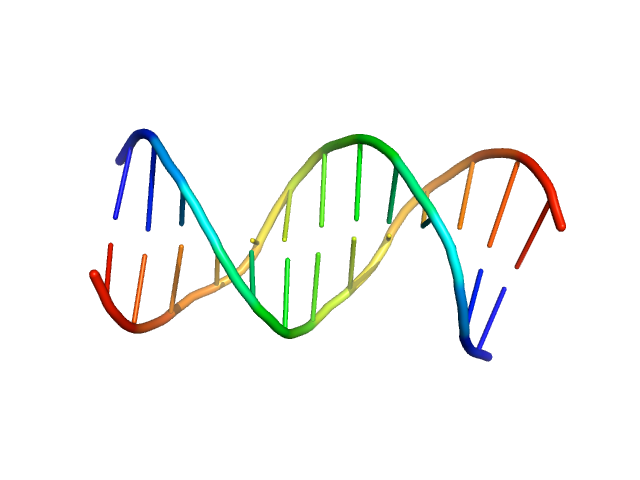
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
13mer dsDNA monomer, 8 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
18 |
nm3 |
|
|