|
|
|
|
|
| Sample: |
Accumulation associated protein (mutant) monomer, 78 kDa Staphylococcus epidermidis (strain … protein
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 1
|
Solution structural studies of pre-amyloid oligomer states of the biofilm protein Aap.
J Mol Biol :167708 (2022)
Yarawsky AE, Hopkins JB, Chatzimagas L, Hub JS, Herr AB
|
| RgGuinier |
14.1 |
nm |
| Dmax |
57.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
Accumulation associated protein (mutant) monomer, 78 kDa Staphylococcus epidermidis (strain … protein
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 1
|
Solution structural studies of pre-amyloid oligomer states of the biofilm protein Aap.
J Mol Biol :167708 (2022)
Yarawsky AE, Hopkins JB, Chatzimagas L, Hub JS, Herr AB
|
| RgGuinier |
14.4 |
nm |
| Dmax |
56.6 |
nm |
|
|
|
|
|
|
|
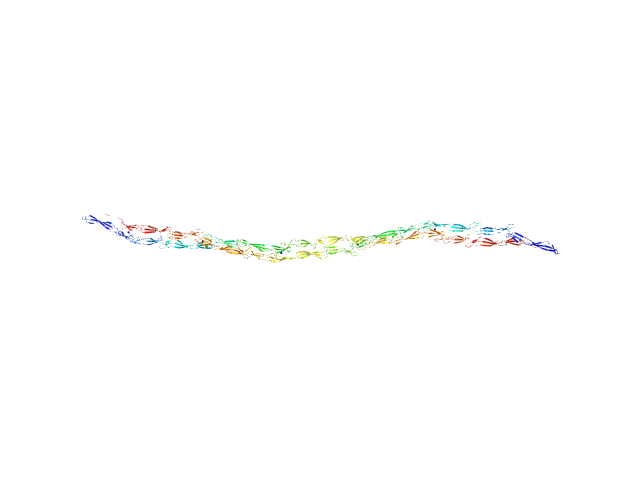
| Sample: |
Accumulation associated protein (mutant) dimer, 155 kDa Staphylococcus epidermidis (strain … protein
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, 5 mM ZnCl2, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 1
|
Solution structural studies of pre-amyloid oligomer states of the biofilm protein Aap.
J Mol Biol :167708 (2022)
Yarawsky AE, Hopkins JB, Chatzimagas L, Hub JS, Herr AB
|
| RgGuinier |
15.0 |
nm |
| Dmax |
63.3 |
nm |
|
|
|
|
|
|
|
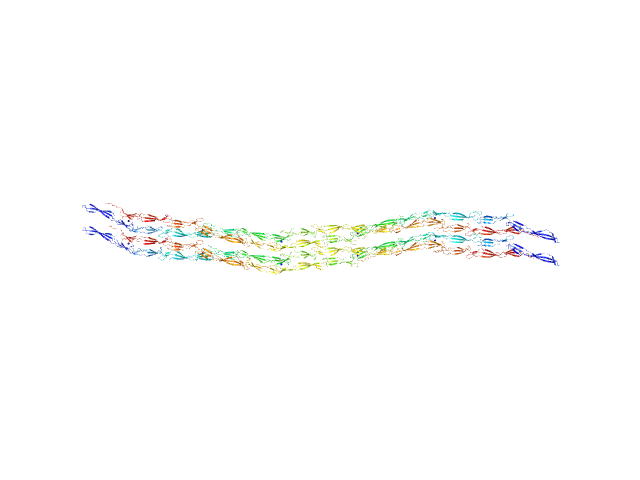
| Sample: |
Accumulation associated protein (mutant) tetramer, 312 kDa Staphylococcus epidermidis (strain … protein
|
| Buffer: |
50 mM MOPS, 50 mM NaCl, 5 mM ZnCl2, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Mar 1
|
Solution structural studies of pre-amyloid oligomer states of the biofilm protein Aap.
J Mol Biol :167708 (2022)
Yarawsky AE, Hopkins JB, Chatzimagas L, Hub JS, Herr AB
|
| RgGuinier |
15.7 |
nm |
| Dmax |
70.0 |
nm |
|
|
|
|
|
|
|
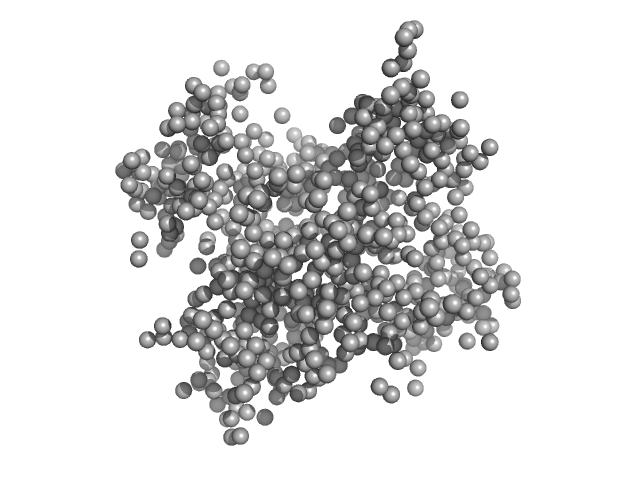
| Sample: |
Piwi domain-containing protein monomer, 91 kDa Pyrococcus furiosus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
|
|
|
|
|
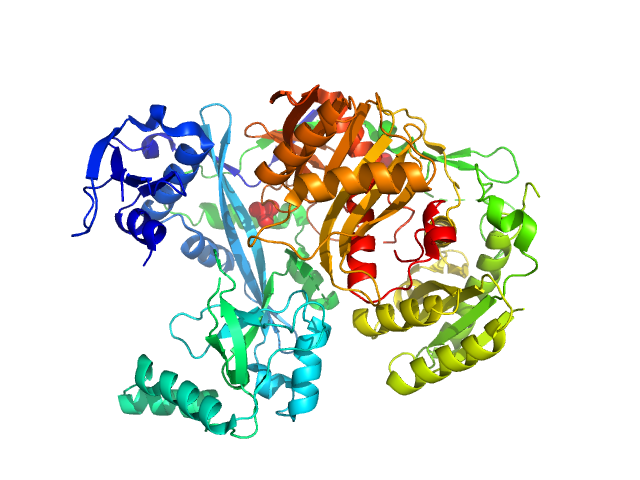
| Sample: |
Piwi domain-containing protein monomer, 91 kDa Pyrococcus furiosus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
|
|
|
|
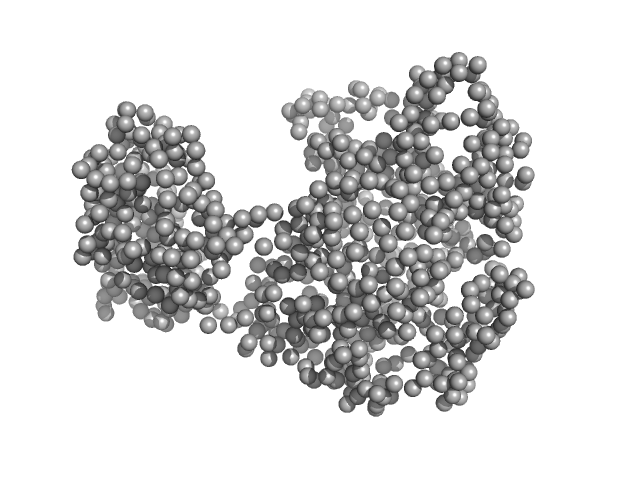
|
| Sample: |
Piwi domain-containing protein monomer, 91 kDa Pyrococcus furiosus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
|
|
|
|
|
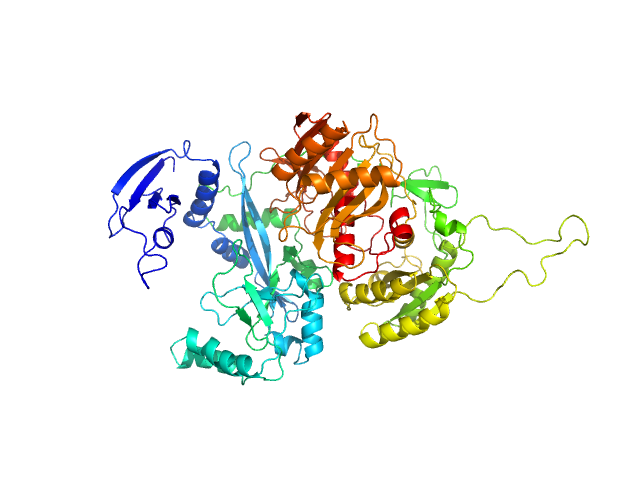
| Sample: |
Piwi domain-containing protein monomer, 91 kDa Pyrococcus furiosus (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 250 mM NaCl, 2 mM DTT, 6 M urea, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Nov 17
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
148 |
nm3 |
|
|
|
|
|
|
|
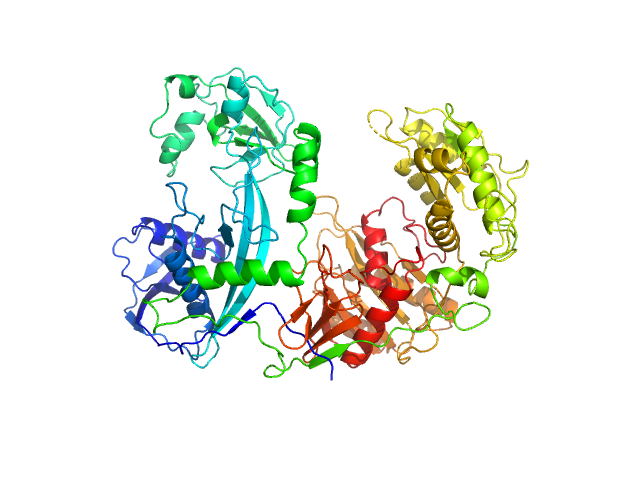
| Sample: |
Clostridium butyricum argonaute protein monomer, 86 kDa protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|
|
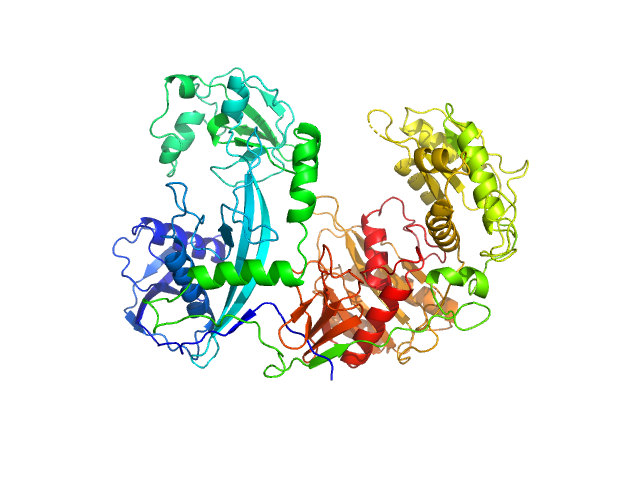
| Sample: |
Clostridium butyricum argonaute protein monomer, 86 kDa protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
185 |
nm3 |
|
|