|
|
|
|
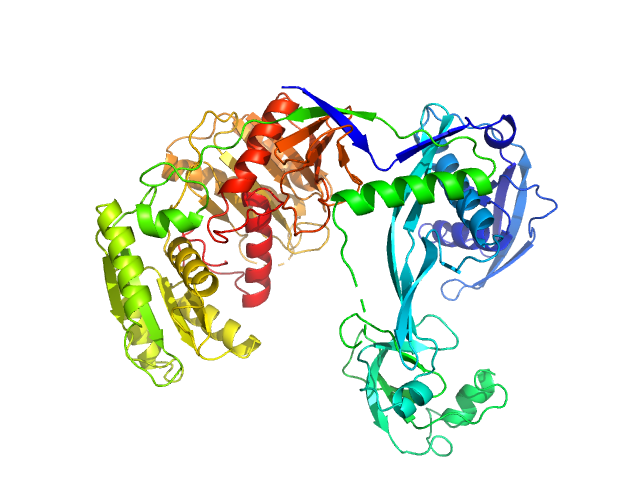
|
| Sample: |
Piwi domain-containing protein monomer, 77 kDa Thermus thermophilus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
|
|
|
|
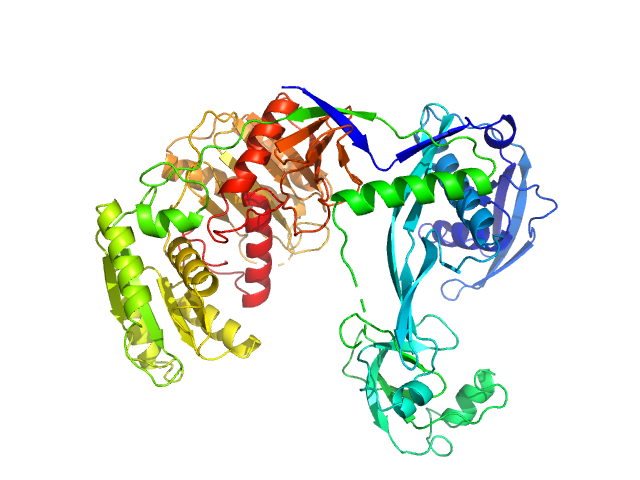
|
| Sample: |
Piwi domain-containing protein monomer, 77 kDa Thermus thermophilus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
|
|
|
|
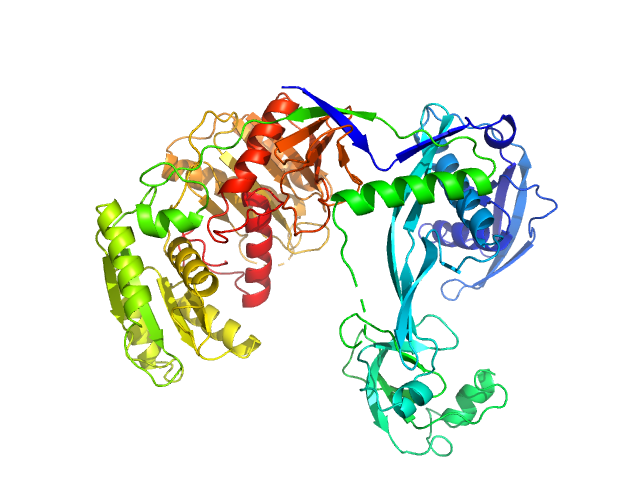
|
| Sample: |
Piwi domain-containing protein monomer, 77 kDa Thermus thermophilus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNA recognition motif (RRM)-containing protein 4 monomer, 111 kDa Ustilago maydis protein
|
| Buffer: |
20 mM Hepes, 200 mM NaCl, 1 mM βME, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 30
|
A MademoiseLLE domain binding platform links the key RNA transporter to endosomes
PLOS Genetics 18(6):e1010269 (2022)
Devan S, Schott-Verdugo S, Müntjes K, Bismar L, Reiners J, Hachani E, Schmitt L, Höppner A, Smits S, Gohlke H, Feldbrügge M, Mitchell A
|
| RgGuinier |
8.8 |
nm |
| Dmax |
30.7 |
nm |
| VolumePorod |
587 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNA recognition motif (RRM)-containing protein 4 NT4 monomer, 40 kDa Ustilago maydis protein
|
| Buffer: |
20 mM Hepes, 200 mM NaCl, 1 mM βME, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 30
|
A MademoiseLLE domain binding platform links the key RNA transporter to endosomes
PLOS Genetics 18(6):e1010269 (2022)
Devan S, Schott-Verdugo S, Müntjes K, Bismar L, Reiners J, Hachani E, Schmitt L, Höppner A, Smits S, Gohlke H, Feldbrügge M, Mitchell A
|
| RgGuinier |
5.6 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mycocerosic acid synthase monomer, 224 kDa Mycobacterium bovis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Oct 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
420 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phenolpthiocerol synthesis type-I polyketide synthase ppsA monomer, 199 kDa Mycobacterium bovis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, 2 mM EDTA, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Jun 14
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
6.7 |
nm |
| Dmax |
20.2 |
nm |
| VolumePorod |
680 |
nm3 |
|
|
|
|
|
|
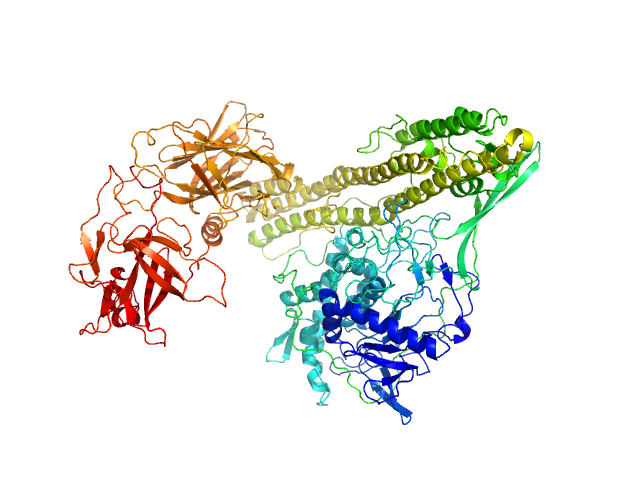
|
| Sample: |
Botulinum Toxin Serotype E, endonegative monomer, 144 kDa Clostridium botulinum protein
|
| Buffer: |
10 mM sodium acetate (20 mM ionic strength), pH: 4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 6
|
Elucidation of critical pH-dependent structural changes in Botulinum Neurotoxin E.
J Struct Biol :107876 (2022)
Lalaurie CJ, Splevins A, Barata TS, Bunting KA, Higazi DR, Zloh M, Spiteri VA, Perkins SJ, Dalby PA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
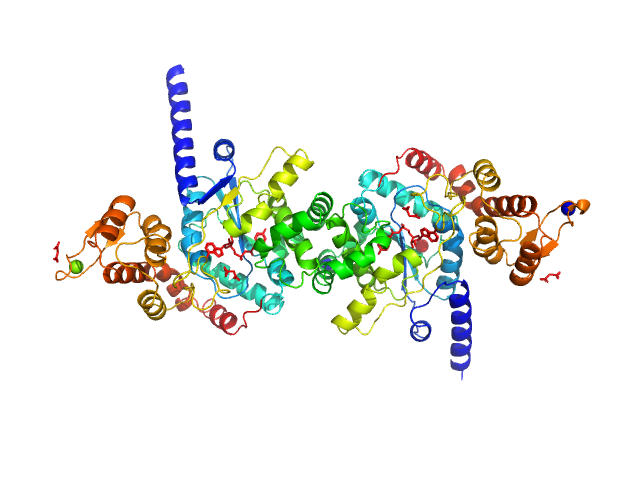
|
| Sample: |
Tyrosine--tRNA ligase dimer, 87 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
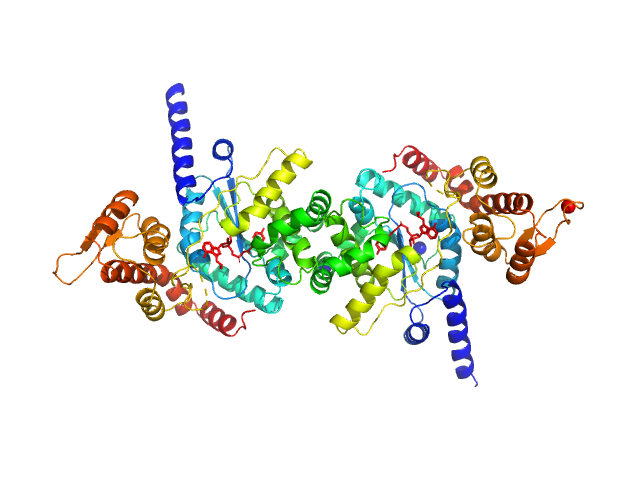
|
| Sample: |
Tyrosine--tRNA ligase (S234C) dimer, 87 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 4
|
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Science 376(6597):1074-1079 (2022)
Xie S, Metcalfe R, Dunn E, Morton C, Huang S, Puhalovich T, Du Y, Wittlin S, Nie S, Luth M, Ma L, Kim M, Pasaje C, Kumpornsin K, Giannangelo C, Houghton F, Churchyard A, Famodimu M, Barry D, Gillett D, Dey S, Kosasih C, Newman W, Niles J, Lee M, Baum J, Ottilie S, Winzeler E, Creek D, Williamson N, Parker M, Brand S, Langston S, Dick L, Griffin M, Gould A, Tilley L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
124 |
nm3 |
|
|