|
|
|
|
|
| Sample: |
DNA polymerase alpha subunit B monomer, 49 kDa Homo sapiens protein
DNA polymerase alpha catalytic subunit monomer, 23 kDa Homo sapiens protein
DNA primase large subunit monomer, 59 kDa Homo sapiens protein
DNA primase small subunit monomer, 50 kDa Homo sapiens protein
29 mer DNA template monomer, 9 kDa DNA
8mer RNA primer monomer, 3 kDa RNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 28
|
Flexibility and distributive synthesis regulate RNA priming and handoff in human DNA polymerase α-primase
Journal of Molecular Biology :168330 (2023)
Cordoba J, Mullins E, Salay L, Eichman B, Chazin W
|
| RgGuinier |
4.9 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
344 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA polymerase alpha subunit B monomer, 49 kDa Homo sapiens protein
DNA polymerase alpha catalytic subunit monomer, 23 kDa Homo sapiens protein
DNA primase small subunit monomer, 50 kDa Homo sapiens protein
DNA primase large subunit monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Apr 15
|
Flexibility and distributive synthesis regulate RNA priming and handoff in human DNA polymerase α-primase
Journal of Molecular Biology :168330 (2023)
Cordoba J, Mullins E, Salay L, Eichman B, Chazin W
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
221 |
nm3 |
|
|
|
|
|
|

|
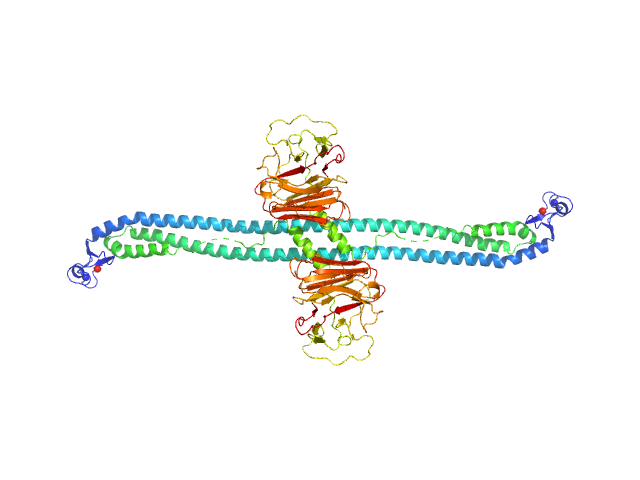
| Sample: |
Tripartite motif-containing protein 72 dimer, 105 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM TCEP, 1 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 7
|
Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat Struct Mol Biol (2023)
Park SH, Han J, Jeong BC, Song JH, Jang SH, Jeong H, Kim BH, Ko YG, Park ZY, Lee KE, Hyun J, Song HK
|
| RgGuinier |
6.9 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
384 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tripartite motif-containing protein 72 dimer, 88 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM TCEP, 1 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Dec 7
|
Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat Struct Mol Biol (2023)
Park SH, Han J, Jeong BC, Song JH, Jang SH, Jeong H, Kim BH, Ko YG, Park ZY, Lee KE, Hyun J, Song HK
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.7 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
|
|
|
|
|
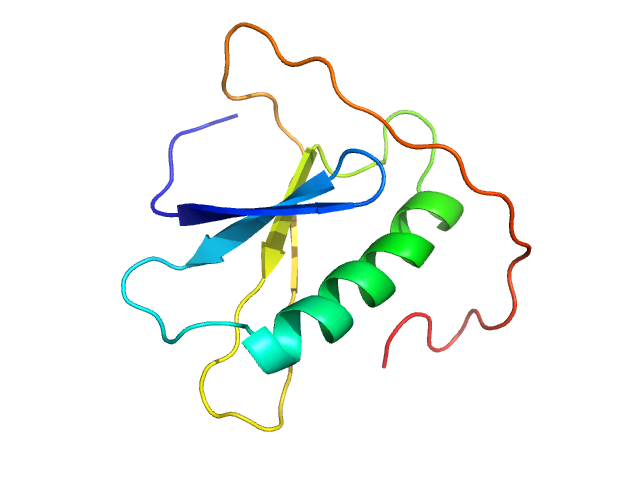
| Sample: |
Cholera toxin transcriptional activator monomer, 12 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, 3% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Vibrio cholerae's ToxRS bile sensing system.
Elife 12 (2023)
Gubensäk N, Sagmeister T, Buhlheller C, Geronimo BD, Wagner GE, Petrowitsch L, Gräwert MA, Rotzinger M, Berger TMI, Schäfer J, Usón I, Reidl J, Sánchez-Murcia PA, Zangger K, Pavkov-Keller T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
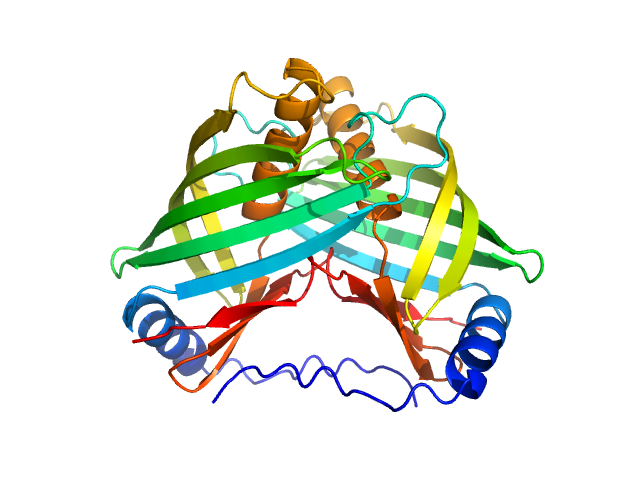
| Sample: |
Transmembrane regulatory protein ToxS dimer, 37 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, 3% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Vibrio cholerae's ToxRS bile sensing system.
Elife 12 (2023)
Gubensäk N, Sagmeister T, Buhlheller C, Geronimo BD, Wagner GE, Petrowitsch L, Gräwert MA, Rotzinger M, Berger TMI, Schäfer J, Usón I, Reidl J, Sánchez-Murcia PA, Zangger K, Pavkov-Keller T
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|
|
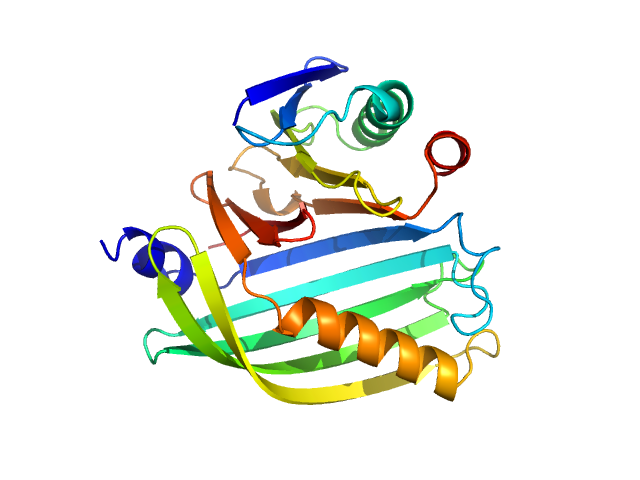
| Sample: |
Cholera toxin transcriptional activator monomer, 12 kDa Vibrio cholerae serotype … protein
Transmembrane regulatory protein ToxS dimer, 37 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, 3% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Vibrio cholerae's ToxRS bile sensing system.
Elife 12 (2023)
Gubensäk N, Sagmeister T, Buhlheller C, Geronimo BD, Wagner GE, Petrowitsch L, Gräwert MA, Rotzinger M, Berger TMI, Schäfer J, Usón I, Reidl J, Sánchez-Murcia PA, Zangger K, Pavkov-Keller T
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|
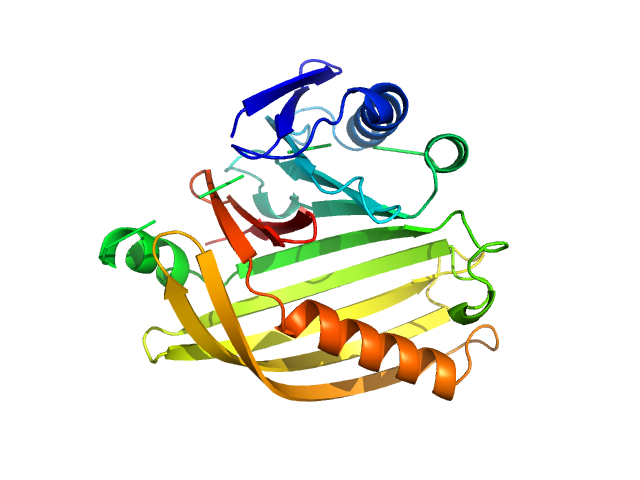
|
| Sample: |
Cholera toxin transcriptional activator monomer, 12 kDa Vibrio cholerae serotype … protein
Transmembrane regulatory protein ToxS dimer, 37 kDa Vibrio cholerae serotype … protein
bile acid: sodium cholate hydrate monomer, 0 kDa
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, 3% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Jul 7
|
Vibrio cholerae's ToxRS bile sensing system.
Elife 12 (2023)
Gubensäk N, Sagmeister T, Buhlheller C, Geronimo BD, Wagner GE, Petrowitsch L, Gräwert MA, Rotzinger M, Berger TMI, Schäfer J, Usón I, Reidl J, Sánchez-Murcia PA, Zangger K, Pavkov-Keller T
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquilin-2 (∆379-462) monomer, 56 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 8
|
Short disordered N-termini & proline-rich domain are major regulators of UBQLN1/2/4 phase separation.
Sarasi Galagedera
|
| RgGuinier |
4.4 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquilin-2 (∆487-538) dimer, 122 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 8
|
Short disordered N-termini & proline-rich domain are major regulators of UBQLN1/2/4 phase separation.
Sarasi Galagedera
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
288 |
nm3 |
|
|