|
|
|
|
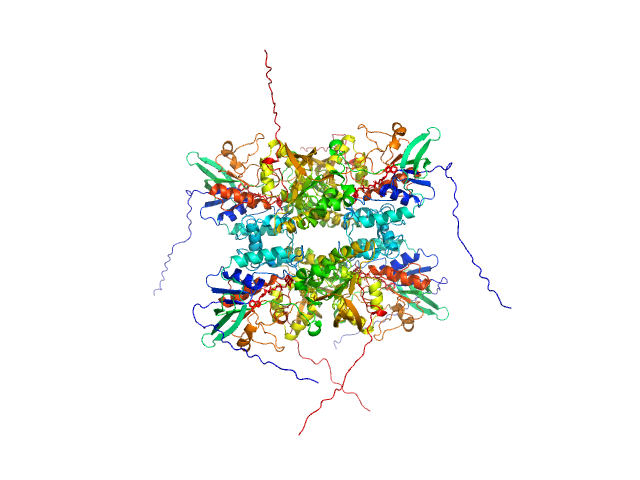
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|
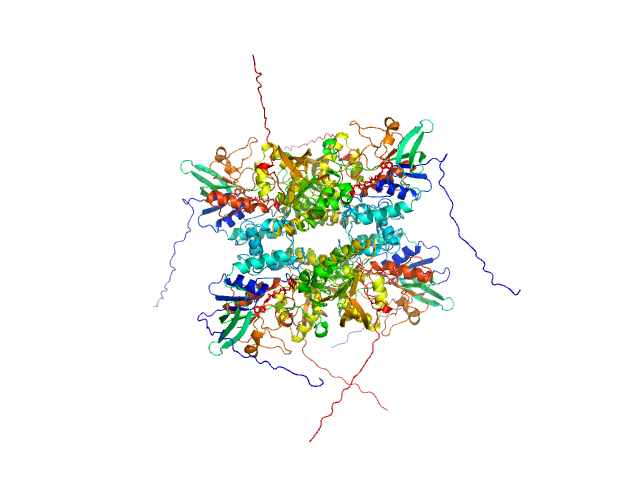
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|
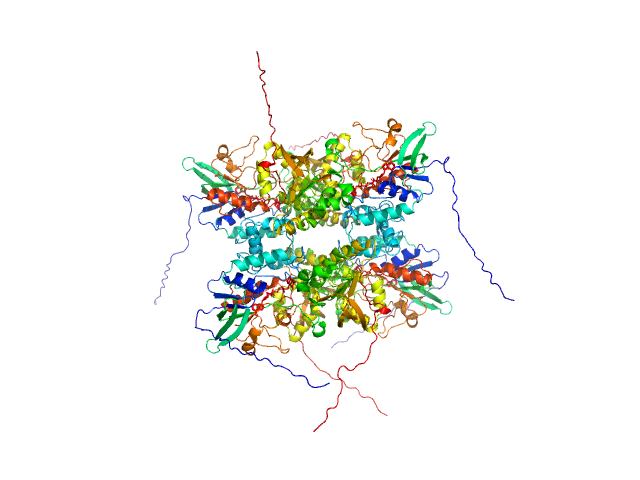
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
390 |
nm3 |
|
|
|
|
|
|
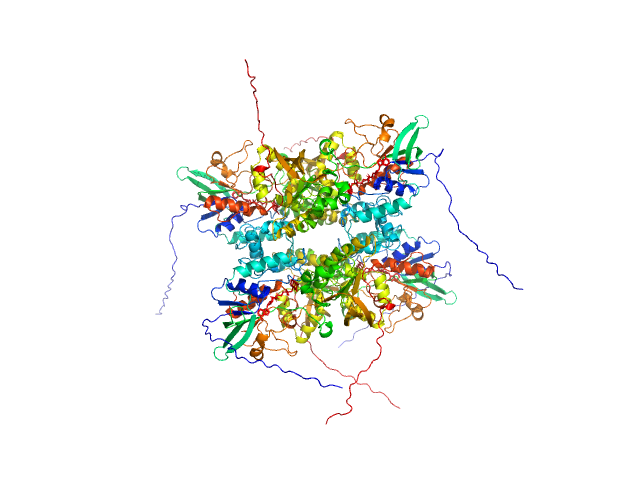
|
| Sample: |
L-lysine 6-monooxygenase (NADPH-requiring) tetramer, 214 kDa Acinetobacter baumannii MRSN … protein
|
| Buffer: |
25 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 14
|
Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N-Hydroxylase from Acinetobacter baumannii.
Biochemistry (2022)
Lyons NS, Bogner AN, Tanner JJ, Sobrado P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
396 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Metacaspase-1 monomer, 46 kDa Candida glabrata (strain … protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1% glycerol, 10 mM CaCl2, pH: 7.6
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 26
|
Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium.
Commun Biol 5(1):1158 (2022)
Conchou L, Doumèche B, Galisson F, Violot S, Dugelay C, Diesis E, Page A, Bienvenu AL, Picot S, Aghajari N, Ballut L
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.4 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Obscurin monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 18
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Obscurin monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Nov 18
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
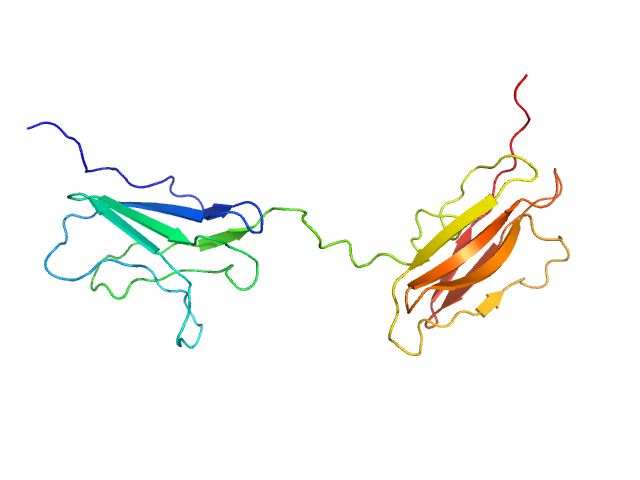
| Sample: |
Obscurin Ig domains 12/13 long monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 20
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
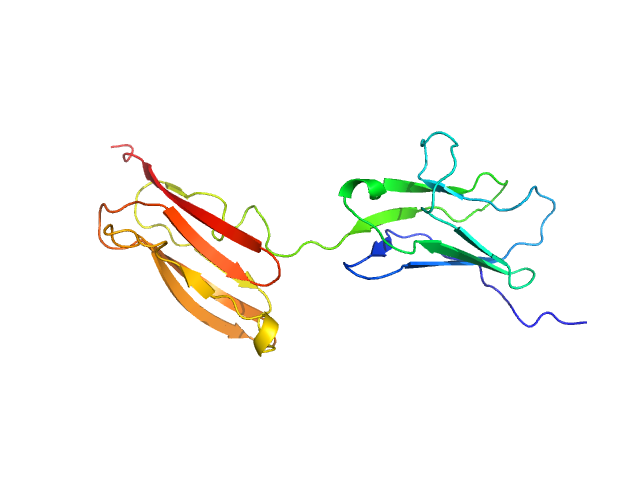
| Sample: |
Obscurin Ig domains 12/13 short monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 20
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
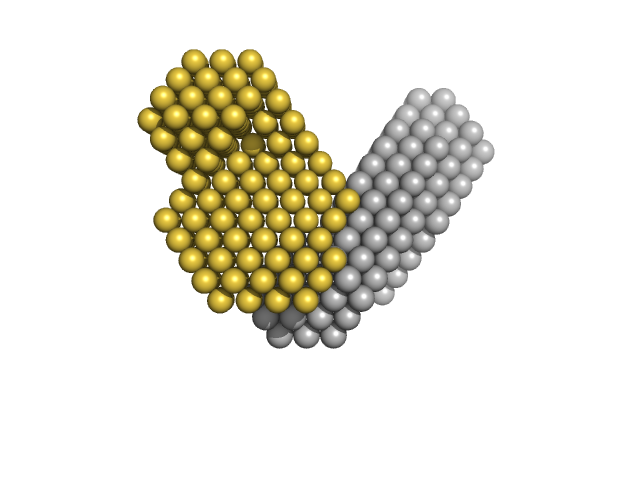
| Sample: |
Ubiquitin-conjugating enzyme E2 D1 (S22R, C85K, D87S) monomer, 17 kDa Homo sapiens protein
Ubiquitin monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8
|
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2019 May 24
|
Production and characterisation of modularly deuterated UBE2D1–Ub conjugate by small angle neutron and X-ray scattering
European Biophysics Journal (2022)
Pietras Z, Duff A, Morad V, Wood K, Jeffries C, Sunnerhagen M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
|
|