|
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 51 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Dec 7
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 104 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2008 Sep 20
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mycocerosic acid synthase monomer, 224 kDa Mycobacterium bovis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 10% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Oct 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
420 |
nm3 |
|
|
|
|
|
|

|
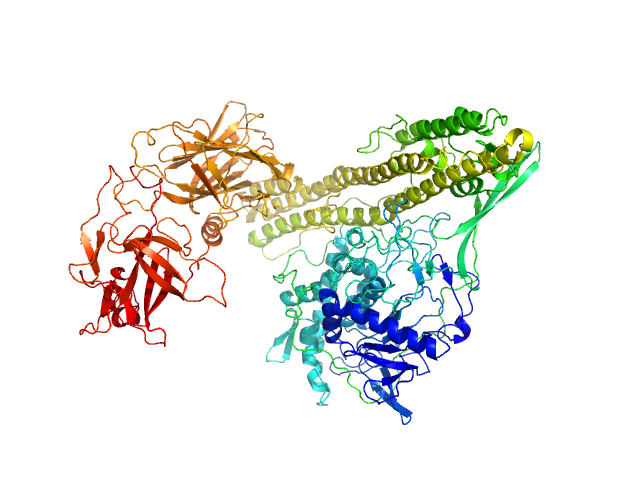
| Sample: |
Phenolpthiocerol synthesis type-I polyketide synthase ppsA monomer, 199 kDa Mycobacterium bovis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, 2 mM EDTA, 10% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Jun 14
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
6.7 |
nm |
| Dmax |
20.2 |
nm |
| VolumePorod |
680 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Botulinum Toxin Serotype E, endonegative monomer, 144 kDa Clostridium botulinum protein
|
| Buffer: |
10 mM sodium acetate (20 mM ionic strength), pH: 4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 6
|
Elucidation of critical pH-dependent structural changes in Botulinum Neurotoxin E.
J Struct Biol :107876 (2022)
Lalaurie CJ, Splevins A, Barata TS, Bunting KA, Higazi DR, Zloh M, Spiteri VA, Perkins SJ, Dalby PA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diacylglycerol kinase alpha monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCl, 200 mM NaCl, 5 mM DTT, 5% v/v glycerol, 3 mM CaCl2, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Dec 8
|
Ca 2+
‐induced structural changes and intramolecular interactions in N‐terminal region of diacylglycerol kinase alpha
Protein Science 31(7) (2022)
Takahashi D, Yonezawa K, Okizaki Y, Caaveiro J, Ueda T, Shimada A, Sakane F, Shimizu N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diacylglycerol kinase alpha monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20mM TrisHCl, 200mM NaCl, 5mM DTT, 5% v/v Glycerol, 3mM EGTA, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Dec 8
|
Ca 2+
‐induced structural changes and intramolecular interactions in N‐terminal region of diacylglycerol kinase alpha
Protein Science 31(7) (2022)
Takahashi D, Yonezawa K, Okizaki Y, Caaveiro J, Ueda T, Shimada A, Sakane F, Shimizu N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
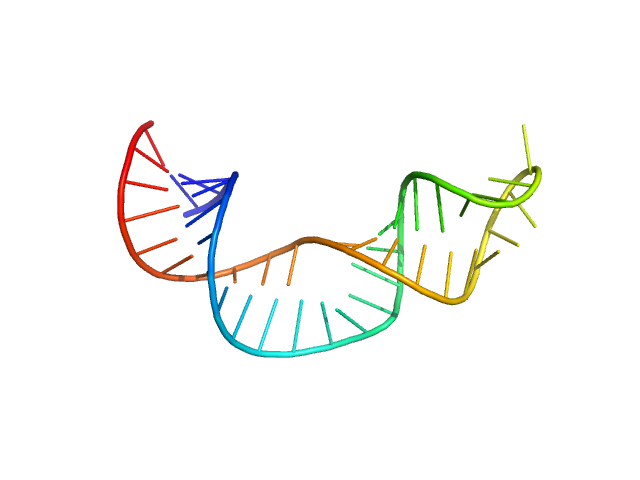
| Sample: |
Non-pre-microRNA stem loop 2 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Solution structure of NPSL2, a regulatory element in the oncomiR-1 RNA.
J Mol Biol :167688 (2022)
Liu Y, Munsayac A, Hall I, Keane SC
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|

|
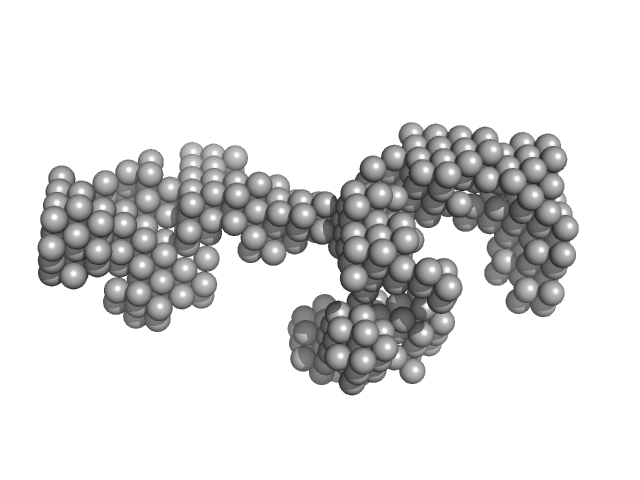
| Sample: |
LincRNA-p21 AluSx1 Sense RNA monomer, 100 kDa Homo sapiens RNA
|
| Buffer: |
50mM HEPES,150 mM NaCl, 15 mM MgCl2, 3% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 1
|
Biophysical characterisation of human LincRNA-p21 sense and antisense Alu inverted repeats.
Nucleic Acids Res 50(10):5881-5898 (2022)
D'Souza MH, Mrozowich T, Badmalia MD, Geeraert M, Frederickson A, Henrickson A, Demeler B, Wolfinger MT, Patel TR
|
| RgGuinier |
6.1 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LincRNA-p21 AluSx1 Antisense RNA monomer, 90 kDa Homo sapiens RNA
|
| Buffer: |
50mM HEPES,150 mM NaCl, 15 mM MgCl2, 3% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 1
|
Biophysical characterisation of human LincRNA-p21 sense and antisense Alu inverted repeats.
Nucleic Acids Res 50(10):5881-5898 (2022)
D'Souza MH, Mrozowich T, Badmalia MD, Geeraert M, Frederickson A, Henrickson A, Demeler B, Wolfinger MT, Patel TR
|
| RgGuinier |
5.9 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
375 |
nm3 |
|
|