|
|
|
|
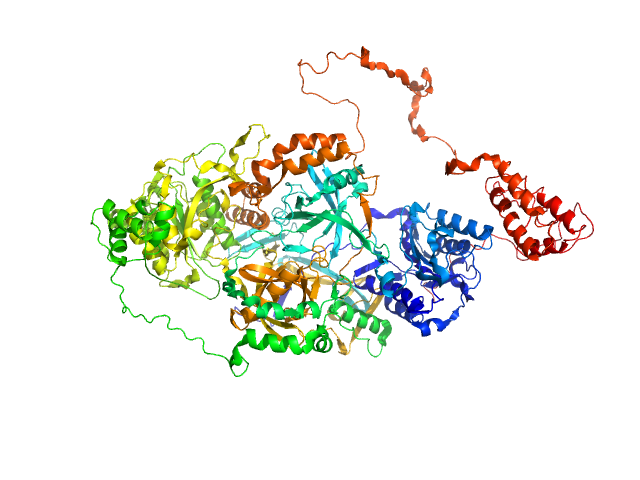
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
|
|
|
|
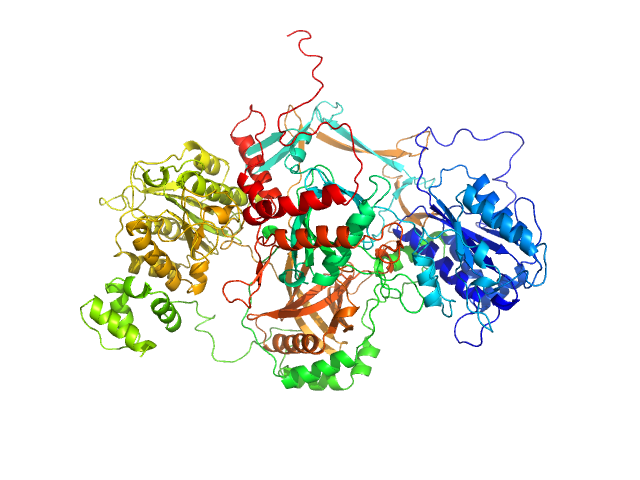
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 ΔCTR monomer, 64 kDa Homo sapiens protein
|
| Buffer: |
50 mM Hepes, 50 mM KCl, 5 mM MgCl2, 5% glycerol and 0.2 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 12
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
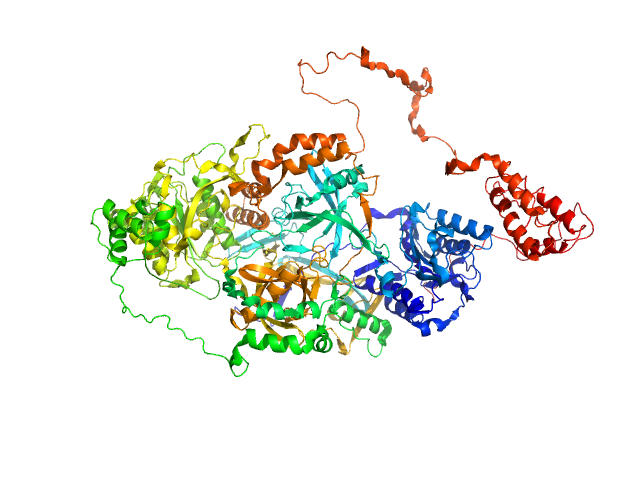
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
Y-DNA monomer, 18 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Jan 8
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
280 |
nm3 |
|
|
|
|
|
|
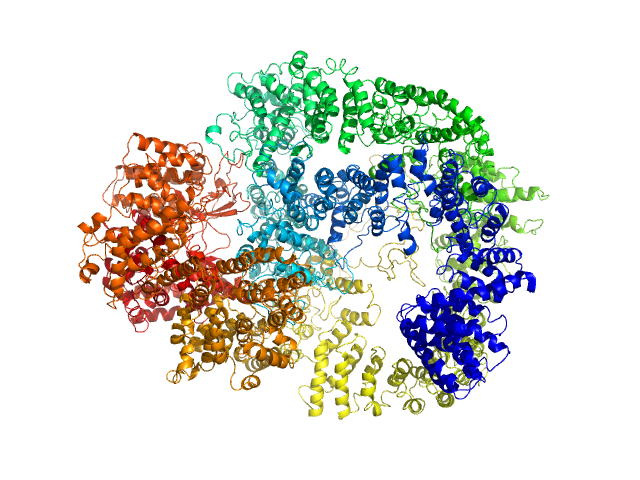
|
| Sample: |
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
5.6 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
1040 |
nm3 |
|
|
|
|
|
|
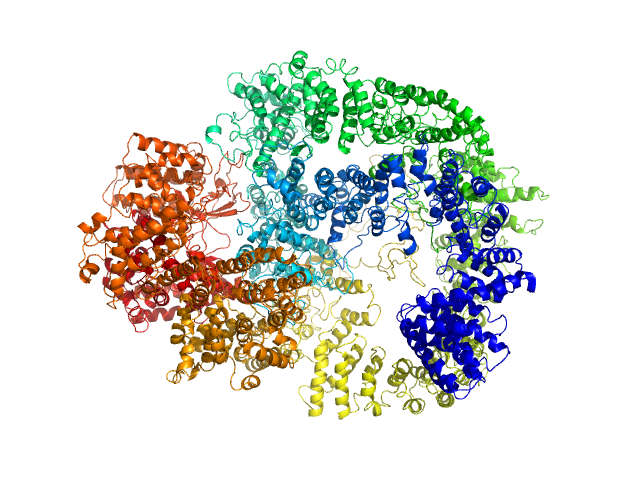
|
| Sample: |
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 100 mM KCl, 5% (v/v) glycerol, 0.2 mM EDTA containing 0.1 mM benzamidine, 0.2 mM PMSF and 0.2 µg/ml pepstatin, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Jan 8
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
5.5 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
918 |
nm3 |
|
|
|
|
|
|
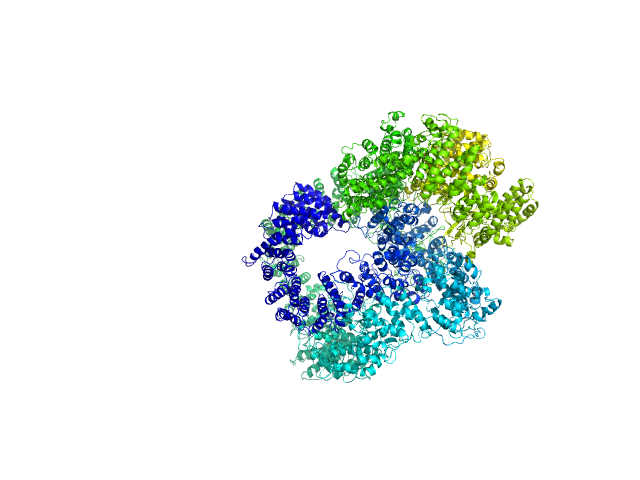
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
dsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
6.5 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
1090 |
nm3 |
|
|
|
|
|
|
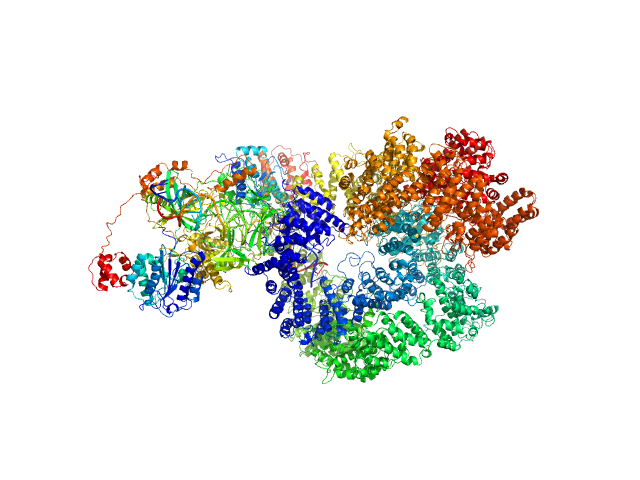
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
dsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
7.5 |
nm |
| Dmax |
29.4 |
nm |
| VolumePorod |
1440 |
nm3 |
|
|
|
|
|
|
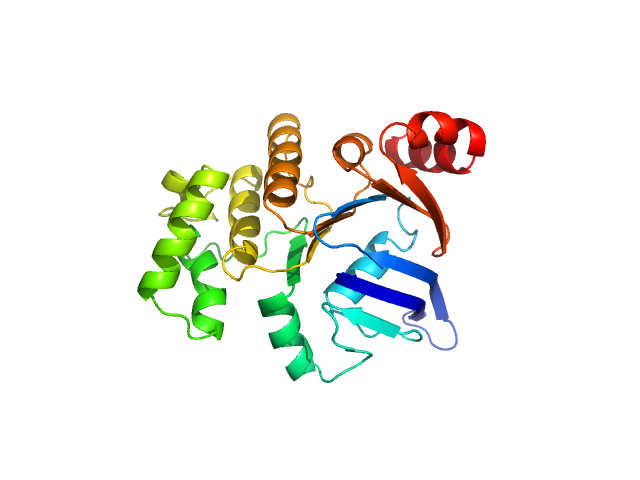
|
| Sample: |
ABC transporter, ATP-binding protein (Nucleotide-Binding Domain SaNsrF) monomer, 31 kDa Streptococcus agalactiae protein
|
| Buffer: |
100 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Dec 11
|
Characterization of the nucleotide-binding domain NsrF from the BceAB-type ABC-transporter NsrFP from the human pathogen Streptococcus agalactiae
Scientific Reports 10(1) (2020)
Furtmann F, Porta N, Hoang D, Reiners J, Schumacher J, Gottstein J, Gohlke H, Smits S
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase monomer, 150 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM DDT, 1 mM EDTA, pH: 8
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Sep 25
|
PfKsgA1 functions as a transcription initiation factor and interacts with the N-terminal region of the mitochondrial RNA polymerase of Plasmodium falciparum.
Int J Parasitol (2020)
Gupta A, Shrivastava D, Shakya AK, Gupta K, Pratap JV, Habib S
|
| RgGuinier |
4.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
295 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase monomer, 150 kDa Saccharomyces cerevisiae protein
rRNA adenine N (6) methyl transferase monomer, 39 kDa Plasmodium falciparum protein
|
| Buffer: |
40 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM DDT, 1 mM EDTA, pH: 8
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2020 Jan 10
|
PfKsgA1 functions as a transcription initiation factor and interacts with the N-terminal region of the mitochondrial RNA polymerase of Plasmodium falciparum.
Int J Parasitol (2020)
Gupta A, Shrivastava D, Shakya AK, Gupta K, Pratap JV, Habib S
|
| RgGuinier |
5.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
393 |
nm3 |
|
|