|
|
|
|
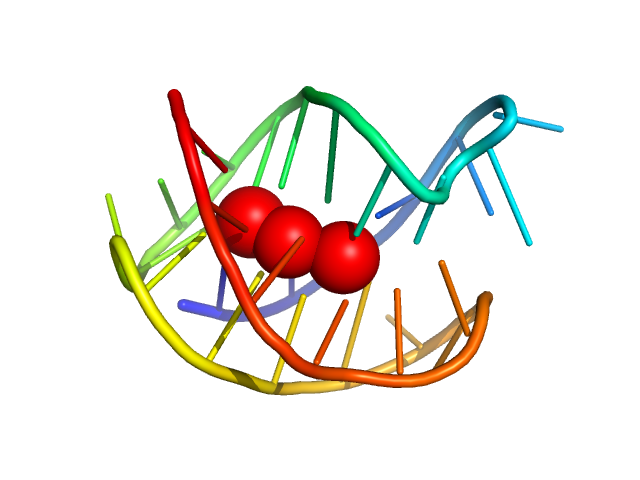
|
| Sample: |
DNA oligonucleotide G4(T4G4)3 monomer, 9 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 12
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.1 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
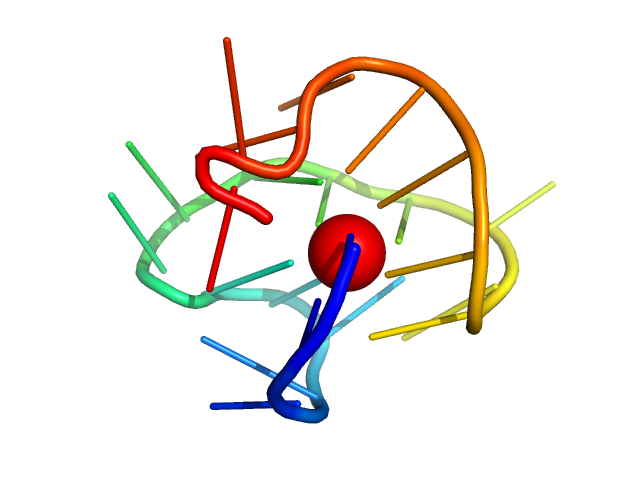
|
| Sample: |
cKIT 2-tetrad promoter GQ antiparallel monomer, 7 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 21
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.0 |
nm |
| Dmax |
3.1 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
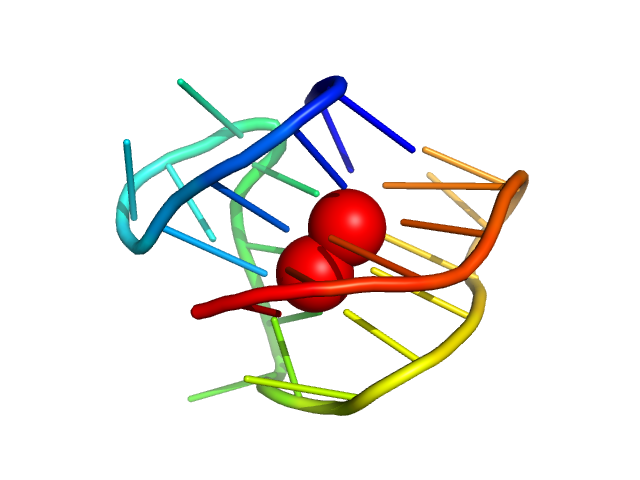
|
| Sample: |
human telomere 24mer hybrid-1 monomer, 8 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 11
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.2 |
nm |
| Dmax |
3.8 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
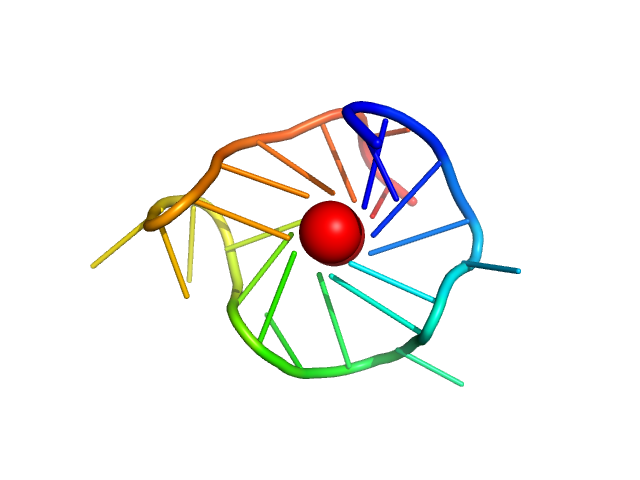
|
| Sample: |
kRas promoter GQ hybrid monomer, 7 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 20
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.9 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
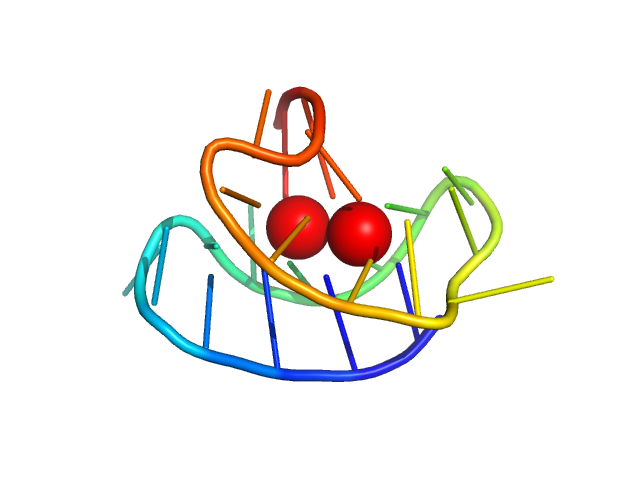
| Sample: |
WNT promoter GQ hybrid form monomer, 7 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 21
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.2 |
nm |
| Dmax |
3.9 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|
|
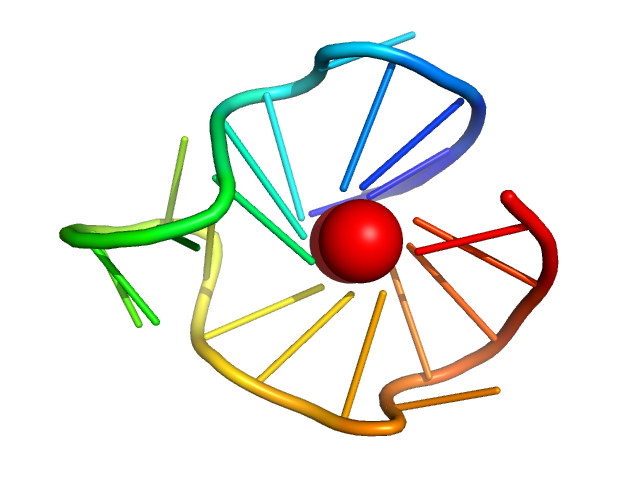
| Sample: |
cKit promoter GQ parallel monomer, 7 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 20
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.6 |
nm |
| Dmax |
3.2 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
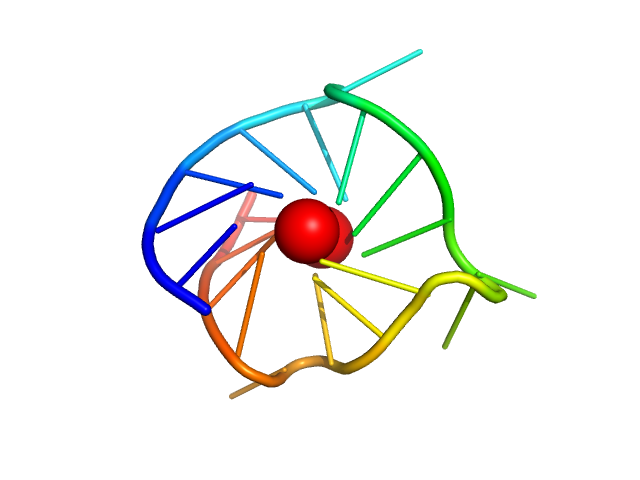
| Sample: |
cMyc promoter GQ parallel monomer, 6 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 20
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.2 |
nm |
| Dmax |
3.9 |
nm |
| VolumePorod |
7 |
nm3 |
|
|
|
|
|
|
|
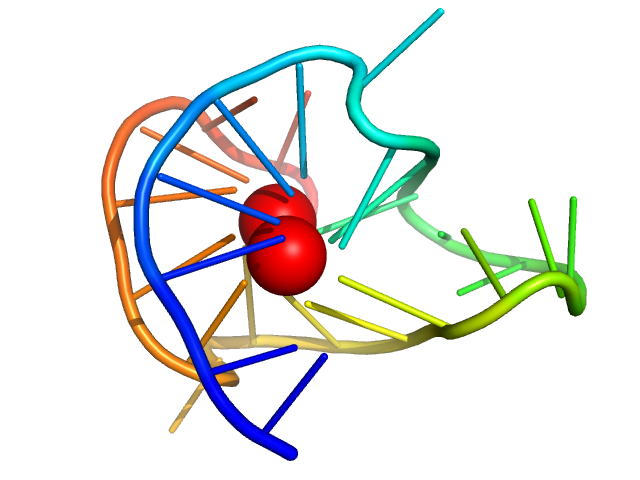
| Sample: |
cMyc promoter GQ 1:6:1 loop parallel monomer, 9 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Nov 20
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
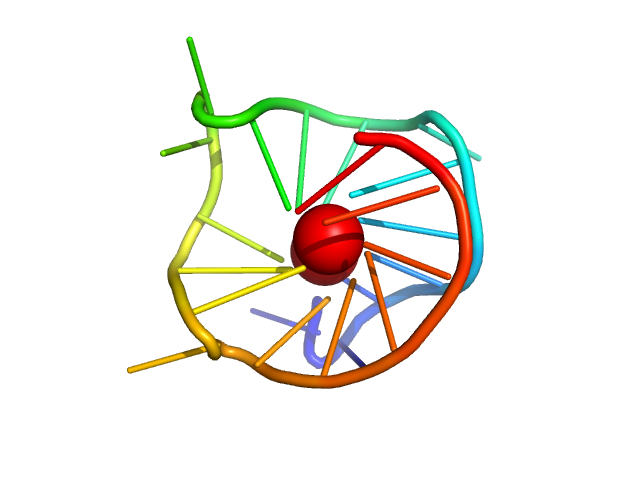
| Sample: |
cMyc promoter modified GQ parallel monomer, 7 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 11
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.1 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|
|
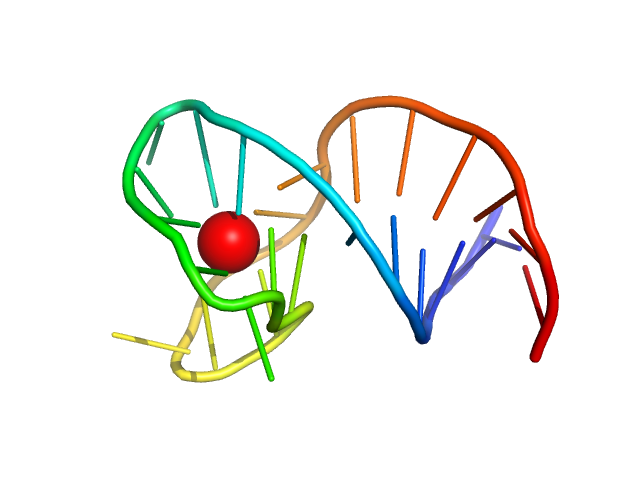
| Sample: |
two-tetrad antiparallel GQ with duplex junction monomer, 10 kDa synthetic construct DNA
|
| Buffer: |
6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, 185 mM KCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 3
|
Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences.
Nucleic Acids Res (2022)
Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.2 |
nm |
| VolumePorod |
12 |
nm3 |
|
|