|
|
|
|
|
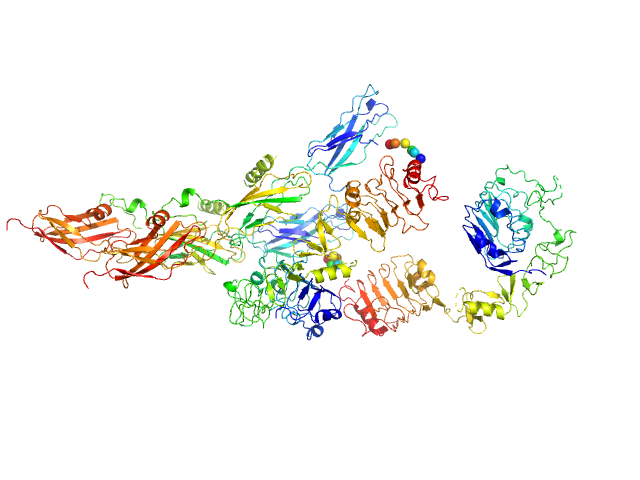
| Sample: |
Cell wall synthesis protein Wag31 , 30 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50mM Tris pH7.5, 300mM NaCl, 10% Glycerol, 1mM EDTA (ethylene diamine tetra acetic acid), 5mM β-mercaptoethanol (BME), pH: 7.5
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2018 Jan 29
|
Higher order assembling of the mycobacterial polar growth factor DivIVA/Wag31.
J Struct Biol :107429 (2019)
Choukate K, Gupta A, Basu B, Virk K, Ganguli M, Chaudhuri B
|
|
|
|
|
|
|
|
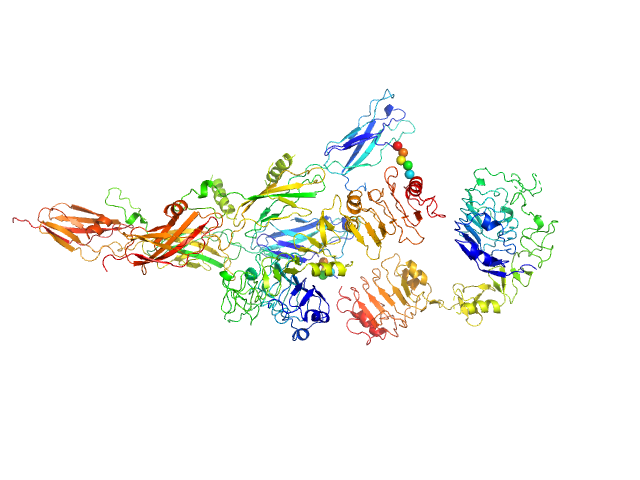
| Sample: |
Insulin receptor-related protein dimer, 201 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 22
|
The dimeric ectodomain of the alkali-sensing insulin receptor-related receptor (ectoIRR) has a droplike shape.
J Biol Chem 294(47):17790-17798 (2019)
Shtykova EV, Petoukhov MV, Mozhaev AA, Deyev IE, Dadinova LA, Loshkarev NA, Goryashchenko AS, Bocharov EV, Jeffries CM, Svergun DI, Batishchev OV, Petrenko AG
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
444 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Insulin receptor-related protein dimer, 201 kDa Homo sapiens protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, pH: 9
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 22
|
The dimeric ectodomain of the alkali-sensing insulin receptor-related receptor (ectoIRR) has a droplike shape.
J Biol Chem 294(47):17790-17798 (2019)
Shtykova EV, Petoukhov MV, Mozhaev AA, Deyev IE, Dadinova LA, Loshkarev NA, Goryashchenko AS, Bocharov EV, Jeffries CM, Svergun DI, Batishchev OV, Petrenko AG
|
| RgGuinier |
5.3 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
430 |
nm3 |
|
|
|
|
|
|
|
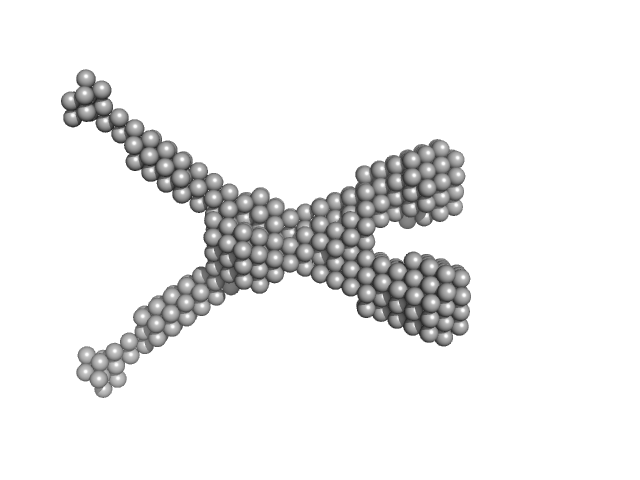
| Sample: |
Noelin tetramer, 72 kDa Mus musculus protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 5
|
Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol 20(1):50 (2019)
Pronker MF, van den Hoek H, Janssen BJC
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 5 , 15 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Feb 14
|
Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
(2019)
Ruiz L, Kaczmarska Z, Gomes T, Aragón E, Torner C, Freier R, Bagiński B, Martin-Malpartida P, de Martin Garrido N, Márquez J, Cordeiro T, Pluta R, Macias M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 8_9 , 15 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Mar 1
|
Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
(2019)
Ruiz L, Kaczmarska Z, Gomes T, Aragón E, Torner C, Freier R, Bagiński B, Martin-Malpartida P, de Martin Garrido N, Márquez J, Cordeiro T, Pluta R, Macias M
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
|
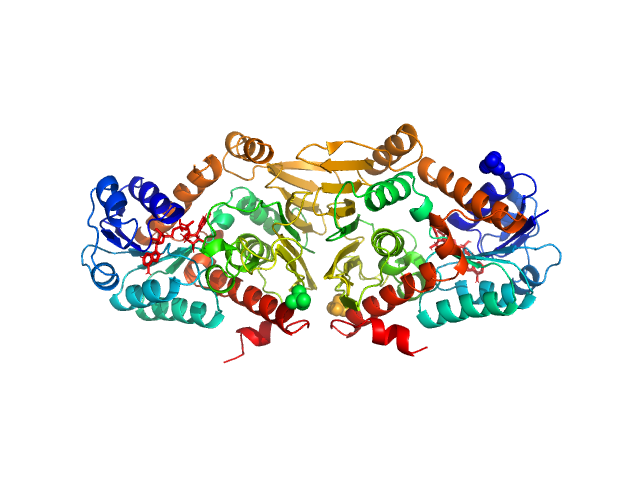
| Sample: |
Escherichia coli YjhC dimer, 86 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1 % (w/v) sodium azide, 5 % (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2018 Apr 12
|
On the structure and function of Escherichia coli YjhC: an oxidoreductase involved in bacterial sialic acid metabolism.
Proteins (2019)
Horne CR, Kind L, Davies JS, Dobson RCJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
C-Jun-amino-terminal kinase-interacting protein 3 dimer, 40 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 0.5 mM TCEP, pH: 7.1
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Sep 22
|
Structural characterization of the RH1-LZI tandem of JIP3/4 highlights RH1 domains as a cytoskeletal motor-binding motif.
Sci Rep 9(1):16036 (2019)
Vilela F, Velours C, Chenon M, Aumont-Nicaise M, Campanacci V, Thureau A, Pylypenko O, Andreani J, Llinas P, Ménétrey J
|
| RgGuinier |
6.3 |
nm |
| Dmax |
23.1 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
C-Jun-amino-terminal kinase-interacting protein 3 dimer, 40 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 0.5 mM TCEP, pH: 7.1
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Sep 22
|
Structural characterization of the RH1-LZI tandem of JIP3/4 highlights RH1 domains as a cytoskeletal motor-binding motif.
Sci Rep 9(1):16036 (2019)
Vilela F, Velours C, Chenon M, Aumont-Nicaise M, Campanacci V, Thureau A, Pylypenko O, Andreani J, Llinas P, Ménétrey J
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.8 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
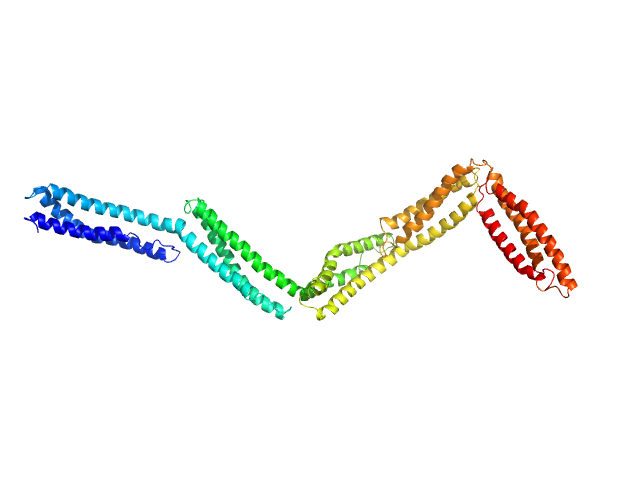
|
| Sample: |
Dystrophin (R11-15 human dystrophin fragment) monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% v/v D2O, pH: 7.1
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 1
|
How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids.
J Struct Biol :107411 (2019)
Mias-Lucquin D, Dos Santos Morais R, Chéron A, Lagarrigue M, Winder SJ, Chenuel T, Pérez J, Appavou MS, Martel A, Alviset G, Le Rumeur E, Combet S, Hubert JF, Delalande O
|
| RgGuinier |
6.2 |
nm |
| Dmax |
27.4 |
nm |
| VolumePorod |
146 |
nm3 |
|
|